Proteins have complex and dynamic shapes. The function of a protein is determined by its structure; a change in the protein’s activity involves a change in some portion of the protein’s structure (shape). What, then, determines a protein’s structure?
Proteins are assembled as a linear chain of amino acids covalently linked by peptide bonds. As this chain is being assembled (each subsequent amino acid is added onto the free carboxy- terminus of the nascent polypeptide chain), the polypeptide chain begins to fold.
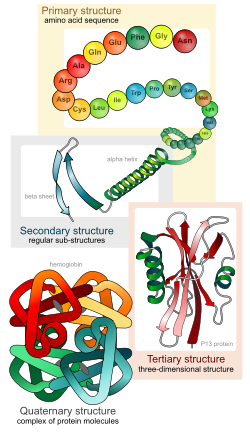
Biologists distinguish 4 levels of protein structure. Students should be able to identify the four levels of protein structure, and the molecular forces or interactions responsible for stabilizing each level of structure.
Primary structure – the linear sequence of amino acids, held together by covalent peptide bonds.
Secondary structure – alpha helices and beta sheets, stabilized by hydrogen bonds between peptide backbone amino groups and carboxyl groups of amino acids within the same polypeptide chain, but not immediately next to each other.

from OpenStax Biology. The dotted lines represent hydrogen bonds. In secondary structures, the hydrogen bonds form between the H- on the amino group and the oxygen atom on the carboxyl group of the peptide backbone. The -R represents amino acid side chains.
Tertiary structure – overall 3-D shape of the folded polypeptide chain, that can be described as the spatial relationships of the secondary structure elements linked by loops. Stabilized by various types of amino acid side chain interactions, including: hydrophobic and van der Waals interactions, hydrogen bonds, ionic bonds, covalent disulfide bonds between cysteine residues, and interactions with solvent water molecules.

From OpenStax Biology. Amino acid side-chain interactions that stabilize protein tertiary structure. Note that tertiary structure involves hydrogen bonds between amino acid side chains, whereas secondary structure involves hydrogen bonds between peptide backbone atoms.
Quaternary structure – assemblage of two or more folded polypeptides into a functional unit. Stabilized by interchain hydrophobic and van der Waals interactions, hydrogen bonding, ionic bonds, and covalent disulfide bonds between cysteine residues on different polypeptide chains. Proteins that function as a single polypeptide chain do not have quarternary structure; only proteins that have two or more polypeptide chains have quarternary structure.
Case examples:
Hemoglobin: The classic case exploring protein structure is hemoglobin. Functional hemoglobin is a tetramer, consisting of two alpha-globin and two beta-globin polypeptide chains. Hemoglobin also requires a cofactor, heme (also called a prosthetic group), containing an iron atom that binds oxygen.
Questions:
a) What levels of protein structure does hemoglobin exhibit?
b) The most common sickle-cell disease mutation changes a glutamic acid (a negatively charged amino acid) in beta-globin to valine (a hydrophobic amino acid). Where would you most commonly expect to find a charged amino acid like glutamic acid, in the interior of the folded protein or on the surface?
c) Which of the following changes do you think might also cause sickle-cell disease?
- the glutamic acid changes to an aspartic acid, a different negatively charged amino acid
- the glutamic acid changes to a lysine, a positively charged amino acid
- the glutamic acid changes to a tryptophan, a hydrophobic amino acid
- the glutamic acid changes to a serine, an uncharged, hydrophilic amino acid
d) Sickle cell hemoglobin mutations alter what levels of protein structure (when sickling of red blood cells is apparent)?
Cystic fibrosis: The most common mutation associated with cystic fibrosis causes a single amino acid, a phenylalanine, to be omitted from the protein called CFTR (cystic fibrosis transmembrane conductor). The CFTR protein functions as a chloride channel in the membrane, formed as the single long CFTR polypeptide chain crosses the membrane back and forth several times. The absence of this phenylalanine, which has a large hydrophobic side chain, causes the protein to be mis-folded. This mis-folded protein is recognized by the cellular quality control system and sent to the cellular recycling center (the proteasome); only about 1 percent makes it to the proper destination, the plasma membrane. My case study is published as a blost post:
Cystic Fibrosis: A Case Study for Membranes and Transport
Extremophiles: Microbes that live in extreme environments of temperature, salt and pH have proteins that are adapted for structural stability in these extreme environments.
This page is adapted from a post originally published on Jung’s Biology Blog https://jchoigt.wordpress.com/2011/10/13/protein-structure-and-function/