Learning Objectives
- Define and use the terms needed to discuss genetic inheritance including gene, allele, dominant, recessive, gamete, genotype, phenotype, homozygote, heterozygote, carrier
- Explain how chromosomal separation at meiosis leads to segregation of alleles in gametes
- Explain how alignment at metaphase results in independent assortment of (unlinked) genes
- Construct and use a Punnett square for a single trait and for two traits using appropriate terminology
- Determine possible offspring types and phenotypic ratios using probability rules
The terminology of Mendelian inheritance
Gregor Mendel is famous for discovering “particulate inheritance” or the idea that hereditary elements are passed on in discrete units rather than “blended” together at each new generation. Today we call those discrete units genes.
- A gene is a hereditary factor that determines (or influences) a particular trait. A gene is comprised of a specific DNA sequence and is located on a specific region of a specific chromosome. Because of its specific location, a gene can also be called a genetic locus.
- An allele is a particular variant of a gene, in the same way that chocolate and vanilla are particular variants of ice cream.
- An organism’s genotype is the particular collection of alleles found in its DNA. An organism with two of the same alleles for a particular gene is homozygous at that locus; an organism with two different alleles for a particular gene is heterozygous at that locus.
- An organism’s phenotype is its observable traits. An organism can have a heterozygous genotype at a particular locus but have a phenotype that looks like only one of the two alleles. This is because one allele is dominant and masks the effect of the other, recessive allele in a dominant/recessive pattern.
- A dominant allele produces its phenotype whether the organism is homozygous or heterozygous at that locus. For example, in humans the allele for brown eyes is dominant to the allele for blue eyes, so a person who is heterozygous at the eye color locus will have brown eyes.
- A recessive allele produces its phenotype only when homozygous at the locus; its phenotype is masked if the locus is heterozygous. For example, a person must have two copies of the blue eye color allele to have blue eyes.
- Sometimes specific recessive alleles are associated with diseases or conditions. A person who is heterozygous for the gene will be phenotypically normal but carry a copy of the recessive, disease-associated allele. This person is said to be a carrier and can pass on the disease allele to his or her offspring.
Crosses with a single trait and the principle of segregation
All of the concepts above are illustrated in the types of experiments that Mendel carried out with pea plants. Pea plants do not sound like a particularly exciting organism to study, but they were very useful in figuring out basic patterns of inheritance! The reason they were so useful is that they have a lot of traits that are caused by a single gene with a simple dominant/recessive inheritance pattern (this is actually pretty rare in general – but more on that later). So what does that statement in bold mean? A classic example is pea color. Peas can be either yellow or green, but not any other colors. Whether they are yellow or green is controlled by a single gene with two alleles, and the yellow allele is dominant to the green allele. The inheritance pattern if you cross homozygous yellow and homozygous green pea plants is illustrated here:
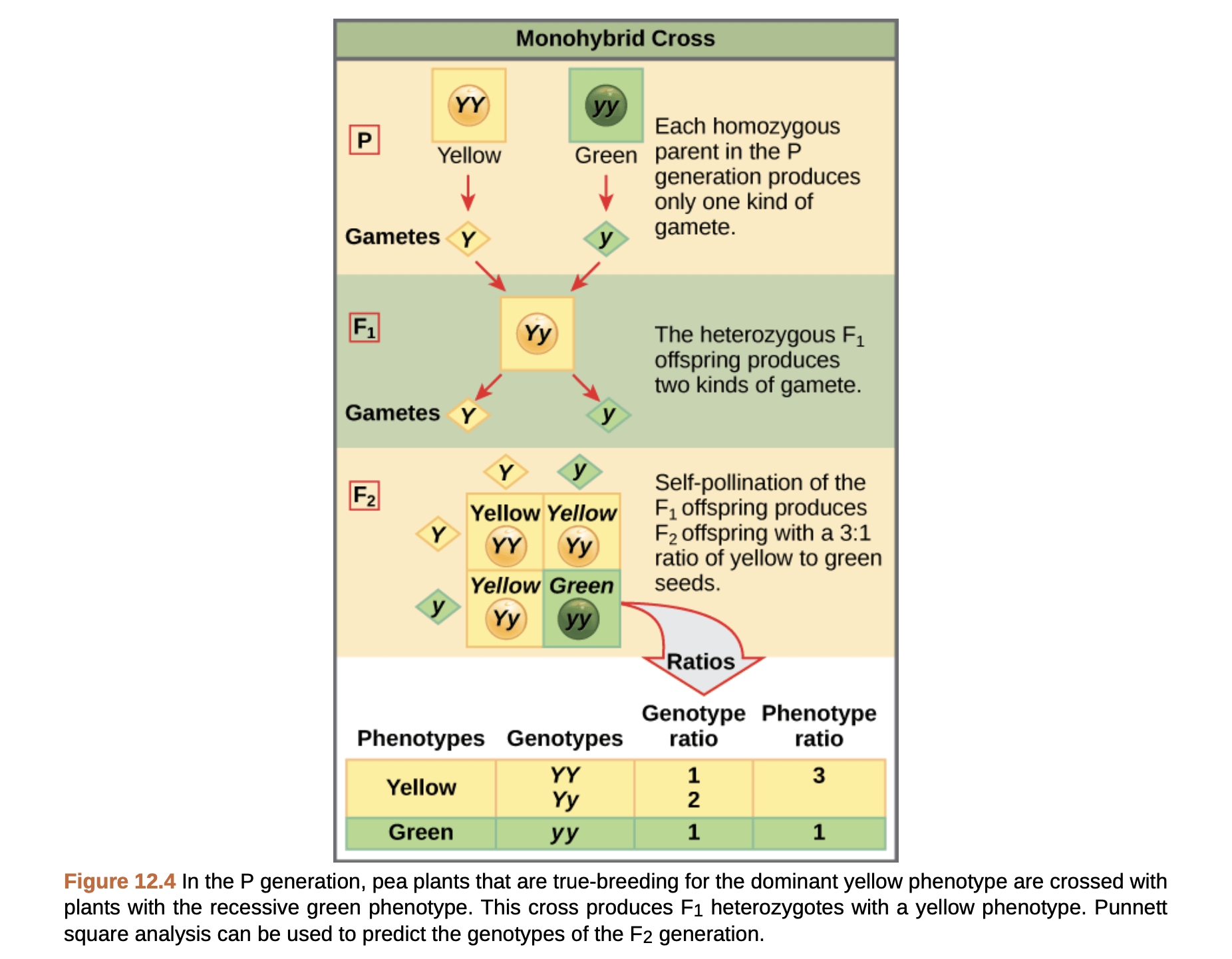
You can see in the first generation (F1) that all offspring produce yellow seeds, even though they have both the yellow and green alleles. If the F1 generation self-fertilizes (pea plants – like most plants – produce both male and female gametes can can mate with themselves), then you now see some offspring that produce yellow peas and some that produce green peas. The yellow:green pea producers exist in approximately a 3:1 ratio, illustrated by constructing a Punnett square.
Punnett squares illustrate the fact that each pea plant gamete contains only one allele for each trait. Even though adult pea plants have two copies of each allele, these two alleles separate into different gametes. Thus when two gametes come together to create a new plant, each gamete carries one allele, resulting in two alleles in the new plant. The idea that each gamete carries only one allele for each trait is the principle of segregation; that is, the two alleles for a particular trait are segregated into different gametes.
Crosses with two traits and the principle of independent assortment
Pea plants have a lot of other traits beyond seed color, and Mendel studied seven other traits. Things become more complex when you follow more than one trait at at time. Here is a cross looking at both pea color (yellow or green) and pea shape (round or wrinkly). Follow the logic of the cross below to see why offspring demonstrate a 9:3:3:1 ratio of different phenotypes.
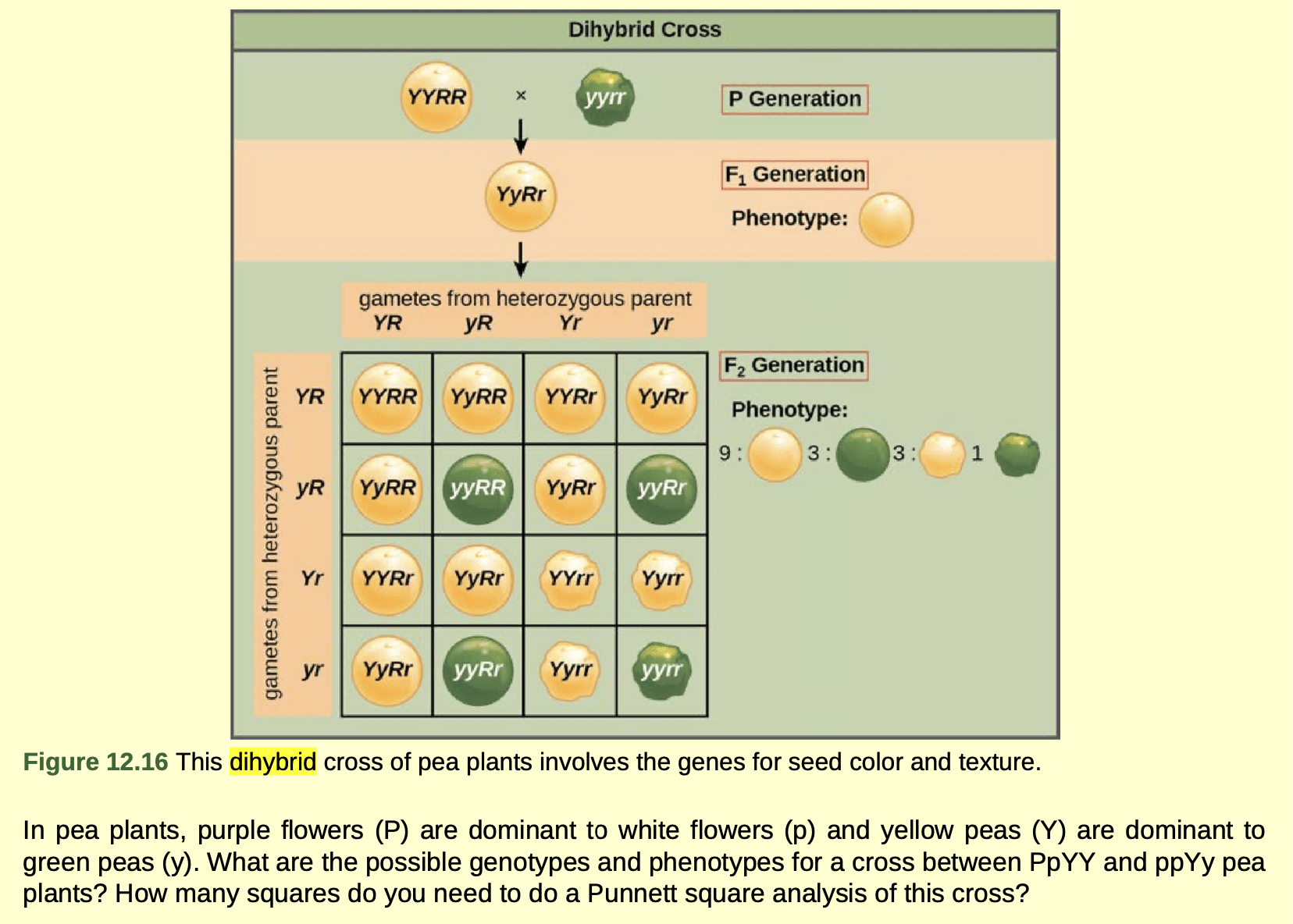
Punnett squares that show two or more traits illustrate the idea that alleles for different traits (different genes) are segregated independently of each other. Yellow seeds are not always round, and green seeds are not always wrinkly; there can be yellow wrinkly seeds, yellow round seeds, green wrinkly seeds, and green round seeds. The idea that alleles for different traits are segregated independently is the principle of independent assortment.
Mendel’s laws and meiosis
Mendel’s laws or principles of segregation and independent assortment are both explained by the physical behavior of chromosomes during meiosis.
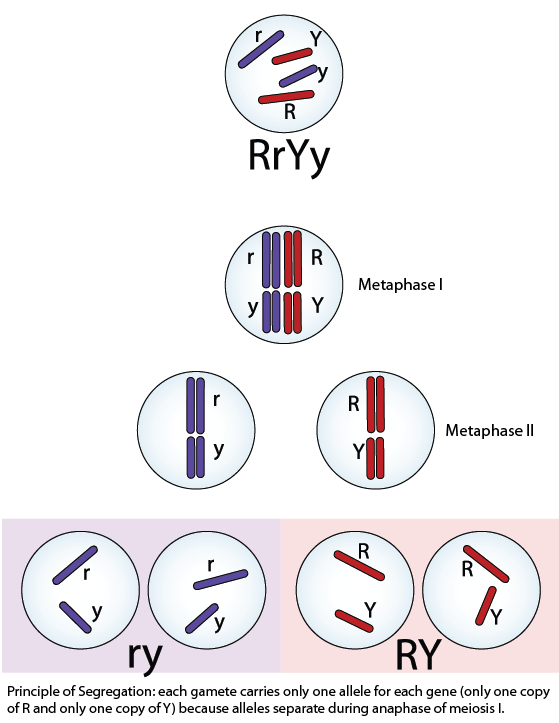
Segregation occurs because each gamete inherits only one copy of each chromosome. Each chromosome has only one copy of each gene; therefore each gamete only contains one or the two possible parental alleles. Segregation occurs when the homologous chromosomes separate during meiotic anaphase I. This principle is illustrated here:
Independent assortment occurs because homologous chromosomes are randomly segregated into different gametes; ie, one gamete does not only get all maternal chromosomes while the other gets all paternal chromosomes. Independent assortment occurs when homologous chromosomes align randomly at the metaphase plate during meiotic metaphase I. This principle is illustrated here:
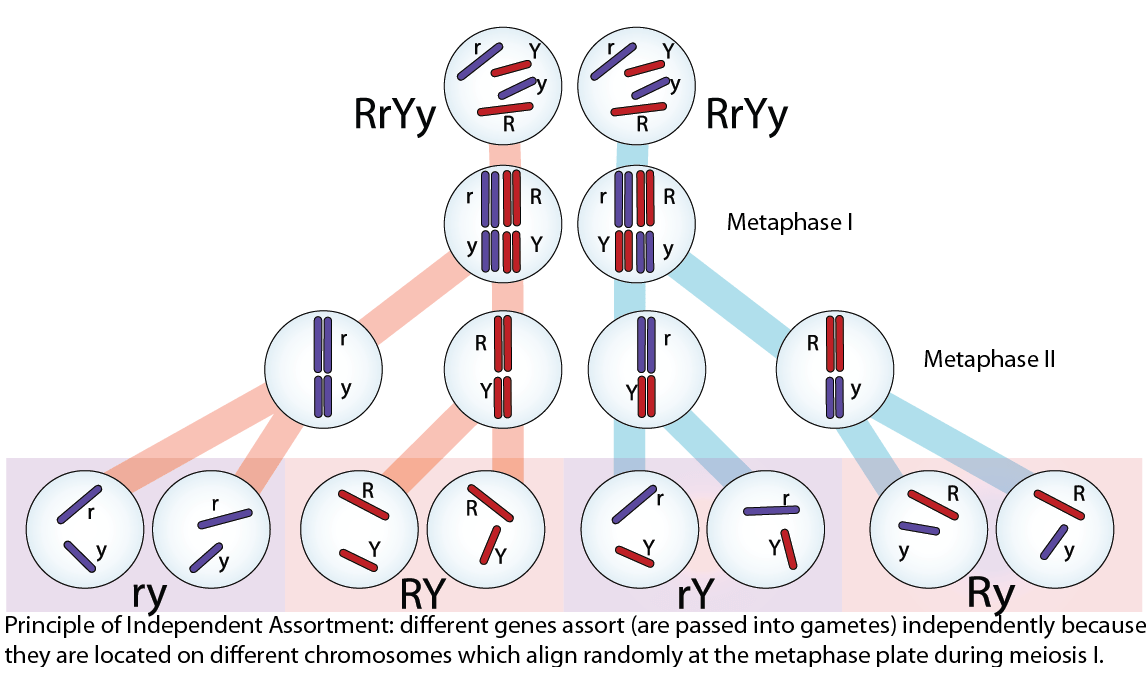
Random, independent assortment during metaphase I can be demonstrated by considering a cell with a set of two chromosomes (n = 2). In this case, there are two possible arrangements at the equatorial plane in metaphase I. The total possible number of different gametes is 2n, where n equals the number of chromosomes in a set. In this example, there are four possible genetic combinations for the gametes. With n = 23 in human cells, there are over 8 million possible combinations of paternal and maternal genotypes in a potential offspring.
Determine possible offspring types and phenotypic ratios using probability rules
Because there can be so many ways for genes to influence the phenotype, in class we will use the information discussed above to determine possible offspring types and phenotypic ratios using simple probability rules. For crosses that involve 2 or more independently assorting traits, using probability rules can be much faster and easier than using 4 x 4 Punnett squares (for 2-factor crosses) or 8 x 8 Punnett squares (for 3-factor crosses). The number of possible gametes is 2N, where N is the number of factors (genes), and the size of the Punnett square needed is 2N x 2N! So instead, we can calculate the outcomes for each factor or gene, then multiply the results.
The two simple probability rules are the product rule and the sum rule
| Product rule | Sum rule |
|---|---|
| For independent events X and Y, the probability (P) of them both occurring (X and Y) is [P(X) x P(Y)]. | For mutually exclusive events X and Y, the probability (P) that one will occur (X or Y) is [P(X) + P(Y)]. |
Example: AaBbCcDd x AaBbCcDd is a cross where A, B, C and D are 4 different genes, with the dominant alleles given as A, B, C, and D, and the recessive alleles are a, b, c, and d, respectively. What proportion of the progeny will have the dominant phenotype for A and B, and recessive for c and d?
If we look at just Aa x Aa, we know that 3/4 of the progeny will have the dominant A phenotype. Similarly, for just Bb x Bb, 3/4 of the progeny will have the dominant B phenotype.
For Cc x Cc, 1/4 of the progeny will have the recessive c phenotype (cc genotype).
For Dd x Dd, 1/4 of the progeny will have the recessive d phenotype (dd genotype).
The rules of probability say that, if these genes are sorting independently, we can just multiply these proportions:
The proportion of ABcd phenotype among the progeny = 3/4 x 3/4 x 1/4 x 1/4 = 9/256
Here’s a quick summary of many of these ideas from Ted Ed:
and here is Khan Academy’s take:
Sustainable Development Goal

UN Sustainable Development Goal (SDG) 3: Good Health and Wellbeing- Understanding genetic inheritance is important for identifying, diagnosing, and treating genetic disorders.

