The standard freshman biology textbook presentation focuses narrowly on glucose metabolism by animal cells, barely touches on fats and amino acids, and ignores most of the metabolic diversity of life. Moreover, the standard textbook version of how this elaborate metabolic network evolved beginning with glycolysis is probably wrong, accordingly to the compelling essay by Lane et al. (2010).
What I prefer is an evolutionary approach that begins with concepts and processes fundamental to all living cells, that must have been present in the last universal common ancestor (LUCA). This page is lightly modified from my original March 2010 blog post: “Evolutionary perspective on energy metabolism”.
Learning objectives:
- Identify what molecule is oxidized, and what molecule is reduced in a redox reaction
- Explain the role of NAD+/NADH as an electron shuttle
- Identify whether an organism is a heterotroph, photoautotroph or chemoautotroph based on their sources of energy and organic carbon
- Explain the difference between substrate-level phosphorylation and oxidative phosphorylation
- Explain how proton gradients are generated across membranes
- Compare and contrast aerobic and anaerobic respiration
- Explain how cells exploit the proton motive force to make ATP
ATP synthesis
We find that all cells – Bacteria, Archaea, Eukarya – use the energy released via ATP hydrolysis (ATP –> ADP + Pi; Pi = inorganic phosphate; deltaG = -7.3 kcal/mol) to perform most of the cellular work. How do cells make ATP? Cells can make ATP in either of two ways: either by substrate-level phosphorylation of ADP, or by oxidative phosphorylation of ADP.
- ATP = adenosine triphosphate
- ADP = adenosine diphosphate
Substrate-level phosphorylation means that a phosphate is transferred to ADP from a high-energy phosphorylated organic compound. A couple of the enzymes in glycolysis make ATP through substrate-level phosphorylation, as well as an enzyme in the citric acid cycle. However, only a small amount of ATP is made this way in cells undergoing respiration.
Oxidative phosphorylation synthesizes the bulk of a cell’s ATP during cellular respiration. A proton-motive force across a membrane provides the energy for ATP synthase (a molecular machine) to make ATP from ADP and inorganic phosphate.
The proton-motive force is created by a large (1000-fold) difference in proton concentrations across a membrane. All prokaryotic cells (bacteria and archaea) maintain a proton gradient (pH gradient) across their plasma membranes. Mitochondria generate a proton gradient across the inner mitochondrial membrane. The interior of the bacterial cell (or the mitochondrial matrix) is relatively alkaline, whereas the exterior periplasmic space (or the mitochondrial intermembrane space) is relatively acidic. The proton motive force arises from a combination of the concentration gradient of protons and a voltage gradient across the membrane (the protons are positively charged, so an imbalance of protons means that the side of the membrane with more protons has a positive charge relative to the side with fewer protons). This proton motive force is a form of stored energy, and protons returning across the membrane down their concentration and voltage gradients release free energy that can be captured by the cell to make ATP. The lipid bilayer membrane is almost impermeable to protons.
This proton gradient (same as a hydrogen ion gradient) is analogous to water stored in an elevated reservoir. The higher the water level in the reservoir, the more potential energy is available to accomplish mechanical work like turning a water wheel to grind grain. In the same way, the greater the difference in proton concentrations across the membrane, the more energy is available for ATP synthase to make ATP. Indeed, the ATP synthase complex even resembles a water wheel, in that the flow of protons down their concentration gradient, through ATP synthase, causes a part of ATP synthase to rotate.
F1ATP Synthase – know this!
The ATP synthase enzyme complex is located in the membrane, and is a remarkable rotor-stator molecular machine (Stock et al. 1999).
The proton motive force drives protons through a channel in the ATP synthase, and turns the rotor at approx 100 rpm. The turning rotor changes the shape of the cytoplasmic subunits (called the F1 ATPase), which bind ADP and inorganic phosphate and bond them together to form ATP. Each 360 degree turn of the rotor results in synthesis of 3 ATP molecules.
The ATP synthases in mitochondria, chloroplasts, and bacteria are all structurally similar, and homologous at the molecular sequence level (Watt et al. 2010). A lesser degree of similarity, and more distant homology, exists with archaeal ATP synthases and vacuolar membrane ATPases that function in active transport of protons across the membrane, using the energy from ATP hydrolysis. Indeed, ATP synthases can work in reverse to hydrolyze ATP and pump protons across the membrane to replenish the membrane proton gradient.
What creates the proton gradient across the membrane, and why is this called “oxidative” phosphorylation?
Chemiosmosis – this is really important!
We have seen how ATP synthase acts like a proton-powered turbine, and uses the energy released from the down-gradient flow of protons to synthesize ATP. The process of pumping protons across the membrane to generate the proton gradient is called chemiosmosis. Chemiosmosis is coupled to the flow of electrons down the electron transport chain, a series of protein complexes in the membrane. Each of these protein complexes accept and pass on electrons, and pump a proton across the membrane for each electron they pass on. Ultimately, the last complex in the electron transport chain passes the electrons to molecular oxygen (O2) to make water, in the case of aerobic respiration.
We define respiration as the passage of electrons down the electron transport chain. We breathe (respire) oxygen because oxygen is the terminal electron acceptor, the end of the line for our mitochondrial electron transport chain. The video below shows the details of the electron transfer reactions, and how they are coupled to pumping protons across the membrane. This is a form of active transport, because the electron transfers release free energy that is used to pump protons against their concentration gradient.
Many bacteria can use other electron acceptors when oxygen is unavailable; we say that they carry on anaerobic respiration, when the electron transport chain functions in the absence of oxygen.
The transfer of electrons from one molecule to another is called an oxidation-reduction, or redox reaction. Oxidative phosphorylation gets its name from oxidation-reduction reactions that occur in the electron transport chain.
Redox Reactions and NAD+/NADH
In cellular metabolism, the most free energy is released by oxidation-reduction reactions, also known as redox reactions. A molecule that loses electrons is oxidized; a molecule that gains electrons is reduced. Different molecules have different tendencies to gain or lose electrons, called the redox potential. A redox reaction between a pair of molecules with a large difference in redox potential results in a large release of free energy. In aqueous environments, the electrons are accompanied by protons. The result is that hydrogen atoms (a proton + electron = hydrogen) are transferred, and many enzymes that carry out redox reactions are called dehydrogenases. Living cells are the original hydrogen fuel cells.
Cellular energy metabolism features a series of redox reactions. Heterotrophs oxidize (take electrons from) organic molecules (food) and give them to (reduce) an electron carrier molecule, called NAD+ (in the oxidized form) that accepts electrons from food to become NADH (the reduced form). NADH then cycles back to NAD+ by giving electrons to (reducing) the first complex of the membrane electron transport chain. Thus NAD+/NADH is a key intermediary in shuttling electrons from food molecules to the electrons transport chain for respiration.
NADH is a high-energy molecule. The oxidation of NADH: NADH + H+ O2 -> NAD+ + H2O is highly exergonic, with a free energy change of -54 kcal/mol (7-fold greater than the free energy change for ATP hydrolysis of -7.3 kcal/mol).
The membrane electron transport chain and chemiosmosis is a strategy for cells to maximize the amount of ATP they can make from the large amounts of free energy available in NADH. The electron transport chain subdivides the oxidation of NADH to a series of lower energy redox reactions, which are used to pump protons across the membrane. The resulting H+ concentration (pH) gradient across the membrane is a form of stored energy, analogous to an electric battery.
Anaerobic respiration
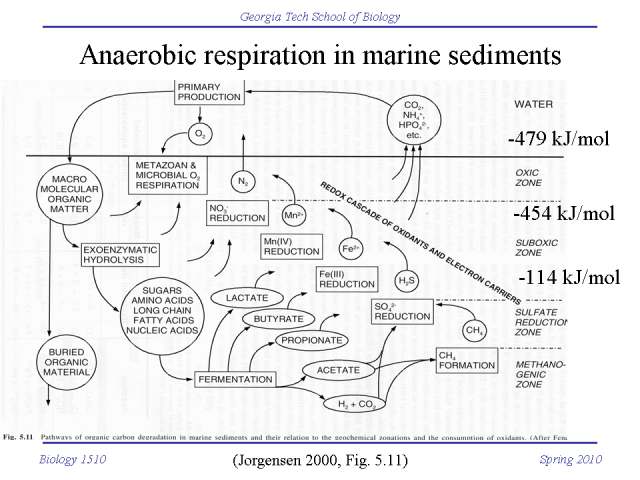
The amount of energy released by these redox reactions, and thus the amount of energy available for ATP synthesis, depends on the redox potential of the terminal electron acceptor. Oxygen (O2) has the greatest redox potential, and thus aerobic respiration results in the most ATP synthesized. Bacteria and Archaea can use other terminal electron acceptors with lower redox potential when oxygen is not available. This anaerobic respiration produces less ATP.
Bacteria can modify their electron transport chains to use a variety of electron donors and electron acceptors, and will switch to the best available electron sources and sinks available in their environment. In marine sediments, microbial communities stratify according to redox potential. The deeper, more anoxic layers use electron acceptors with progressively lower reducing potential.
In addition to ATP synthesis, prokaryotic cells can use the proton motive force to supply energy for active transport of molecules across the plasma membrane, and to power the motor complex that rotates the bacterial flagellum. In eukaryotes, mitochondria maintain a proton gradient across the inner mitochondrial membrane and chloroplasts maintain a proton gradient across the thylakoid membrane. Since both these organelles are thought to have originated as prokaryotic endosymbionts, the mitochondrial inner membrane and the thylakoid membrane correspond to the original plasma membranes of the ancestral endosymbionts. Both mitochondria and chloroplasts use the proton gradient for ATP synthesis and active transport of metabolites into and out of the organelles.
Oxidative phosphorylation coupled to redox pathways that release large amounts of free energy from food molecules, and makes the vast majority of the ATP in respiring cells. Surprisingly, oxidative phosphorylation appears to be at least as ancient as glycolysis and fermentation, and operates in both anaerobic and aerobic environments. Recall that “oxidative” refers not to oxygen gas, but to oxidation-reduction reactions. Oxidative phosphorylation gets the energy required for ATP synthesis from the transfer of electrons from a higher energy electron donor to a lower energy electron acceptor via the membrane electron transport chain. If life originated in alkaline hydrothermal vent environments (Lane et al. 2010) such as the Lost City pictured below, the reducing environment would have provided abundant high-energy electron donors for ATP synthesis by oxidative phosphorylation.

Cellular Metabolic Pathways
The energy for ATP synthesis via either substrate-level phosphorylation or oxidative phosphorylation comes from organic molecules (such as carbohydrates), or from sunlight, or from inorganic electron donors. We can classify organisms according to their source of energy and organic carbon:
- heterotrophs – get energy and organic carbon from metabolism of pre-existing organic compounds (food)
- photoautotrophs – use energy from sunlight to make their own organic carbon compounds from carbon dioxide
- chemoautotrophs – use energy from inorganic chemicals to make their own organic carbon compounds from carbon dioxide
Metabolic pathways carry out reactions that capture energy from these various sources (organic compounds, sunlight or chemicals) and couple them to synthesis of ATP from ADP.
An evolutionary perspective
The generation of a proton gradient across a membrane and chemiosmosis are universal to life on earth, and are fundamental ways for cells to make a living. Lane and colleagues speculate that “proton power” may have been the earliest form of energy metabolism, essential to, and pre-dating, the last universal common ancestor, LUCA (Lane, 2009; Lane et al. 2010).
The earliest cells, living in anoxic environments, used various electron acceptors. After cyanobacteria caused the Great Oxygenation Event, bacteria that adapted their electron transport chains to exploit oxygen as the terminal electron acceptor gained higher energy yield and thus a tremendous selective advantage. One line of aerobic bacteria took up an endosymbiotic relationship within a larger host cell, providing ATP in exchange for organic molecules. This symbiosis must have occurred in the ancestor of all eukaryotes, because all existing eukaryotes have mitochondria (Martin and Mentel, 2010). The evidence for the endosymbiont origin of mitochondria can be found in:
- the double membrane of mitochondria, where the inner membrane contains the electron transport chain, just as the plasma membrane of the aerobic endosymbiont
- the DNA of mitochondria, whose circular chromosome and genetic sequences resemble the alpha-proteo bacteria.
- the ribosomes of mitochondria, that resemble prokaryotic ribosomes
Powered by hundreds or even thousands of mitochondria, eukaryotic cells attained larger sizes and evolved true multicellular lifestyles.
My video on this topic, 23min. (includes the 3 short segments excerpted above)
Powerpoint slides used in the video above:
B1510_module3_5_respiration_2011Fall
Put it all together: Microbial fuel cells
References:
Lane, N 2009 Was our oldest ancestor a proton-powered rock? New Scientist 19 October 2009 https://www.newscientist.com/article/mg20427306.200-was-our-oldest-ancestor-a-protonpowered-rock.html?page=1
Lane, N, JF Allen, W Martin 2010 How did LUCA make a living? Chemiosmosis in the origin of life. BioEssays DOI 10.1002/bies.200900131
Stock D, Leslie AGW, Walker JE 1999 Molecular architecture of the rotary motor in ATP synthase. Science 286:1700–1705. Abstract/FREE Full Text
Watt, IN, MG Montgomery, MJ Runswick, AGW Leslie, JE Walker 2010 Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria PNAS 107 : 16823-16827 doi:10.1073/pnas.1012260107
The standard freshman biology textbook presentation focuses narrowly on glucose metabolism by animal cells, barely touches on fats and amino acids, and ignores most of the metabolic diversity of life. Moreover, the standard textbook version of how this elaborate metabolic network evolved beginning with glycolysis is probably wrong, according to the compelling essay by Lane et al. (2010).
I prefer an evolutionary approach that begins with concepts and processes that are fundamental to all living cells, that must have been present in the last universal common ancestor (LUCA). This page is lightly modified from my original March 2010 blog post: “Evolutionary perspective on energy metabolism”.
Learning objectives:
- Identify what molecule is oxidized, and what molecule is reduced in a redox reaction
- Explain the role of NAD+/NADH as an electron shuttle
- Identify whether an organism is a heterotroph, photoautotroph or chemoautotroph based on their sources of energy and organic carbon
- Explain the difference between substrate-level phosphorylation and oxidative phosphorylation
- Explain how proton gradients are generated across membranes
- Compare and contrast aerobic and anaerobic respiration
- Explain how cells exploit the proton motive force to make ATP
- Hypothesize about how the earliest cells could make ATP in the absence of oxygen
How do cells get the energy to perform work?
ATP synthesis
We find that all cells – Bacteria, Archaea, Eukarya – use the energy released via ATP hydrolysis to perform most of their work. How do cells make ATP? Cells can regenerate ATP from ADP + inorganic phosphate in either of two ways: either by substrate-level phosphorylation, or by oxidative phosphorylation.
Substrate-level phosphorylation means that a phosphate is transferred to ADP from another phosphorylated organic compound. A couple of the enzymes in glycolysis make ATP through substrate-level phosphorylation, as well as an enzyme in the citric acid cycle. However, these reactions tap only a small fraction of the potential energy in glucose, so that only a small amount of ATP is made this way in cells undergoing respiration. Substrate-level phosphorylation by glycolysis enzymes is the major source of ATP only in cells undergoing fermentation (fermentative cells perform glycolysis, but no respiration).
Oxidative phosphorylation is an apparently more complex process where a proton-motive force across a membrane powers an ATP synthase enzyme complex (a molecular machine) to make ATP from ADP and inorganic phosphate. Oxidative phosphorylation is coupled to redox pathways that release large amounts of free energy from food molecules, and makes the vast majority of the ATP in respiring cells. Surprisingly, oxidative phosphorylation appears to be at least as ancient as glycolysis and fermentation, and operates in both anaerobic and aerobic environments. The “oxidative” refers not to oxygen gas, but to oxidation-reduction reactions. Oxidative phosphorylation gets the energy required for ATP synthesis from the transfer of electrons from a higher energy electron donor to a lower energy electron acceptor. If life originated in alkaline hydrothermal vent environments (Lane et al. 2010) such as the Lost City pictured below, the reducing environment would have provided abundant high-energy electron donors for ATP synthesis by oxidative phosphorylation.

Cellular Metabolic Pathways
The energy for ATP synthesis via either substrate-level phosphorylation or oxidative phosphorylation comes from organic molecules (such as carbohydrates), or from sunlight, or from inorganic electron donors. We can classify organisms according to their source of energy and organic carbon:
- heterotrophs – get energy and organic carbon from metabolism of pre-existing organic compounds (food)
- photoautotrophs – use energy from sunlight to make their own organic carbon compounds from carbon dioxide
- chemoautotrophs – use energy from inorganic chemicals to make their own organic carbon compounds from carbon dioxide
Metabolic pathways carry out reactions that capture energy from these various sources (organic compounds, sunlight or chemicals) and couple them to synthesis of ATP from ADP.
Redox Reactions
In these metabolic pathways, we find that the most free energy is released by oxidation-reduction reactions, also known as redox reactions. Redox reactions are electron transfer reactions. A molecule that loses electrons is oxidized; a molecule that gains electrons is reduced. Different molecules have different tendencies to gain electrons, called the redox potential. A redox reaction between a pair of molecules with a large difference in redox potential results in a large release of free energy. In cells and aqueous environments, the electrons are accompanied by protons (protons are always present in water, from dissociation of water molecules to H+ and OH-). The result is that hydrogen atoms are transferred, and many enzymes that carry out redox reactions are called dehydrogenases. Living cells are the original hydrogen fuel cells!
Cellular energy metabolism features a series of redox reactions. Heterotrophs oxidize (take electrons from) organic molecules (food) and give them to an electron carrier molecule, called NAD+ (in the oxidized form) that accepts electrons from food to become NADH (the reduced form). NADH then cycles back to NAD+ by giving electrons to (reducing) an electron acceptor protein in a membrane.
NADH is a high-energy molecule. The oxidation of NADH: NADH + H+ O2 -> NAD+ + H2O is highly exergonic, with a free energy change of -54 kcal/mol. In the membrane, the electrons are transferred down an electron transport chain, consisting of a series of membrane proteins and molecules with increasing redox potential. Components of the electron transport chain use the sequential releases of free energy to pump protons across the membrane against their electrochemical gradient. The resulting H+ concentration (pH) gradient across the membrane is a form of stored energy, analogous to an electric battery. At the end of the electron transport chain is the terminal electron acceptor. The terminal electron acceptor is molecular oxygen (O2) in aerobic respiration, and other molecules such as nitrate, iron, or sulfate in anaerobic respiration. The video below shows the electron transport chain in mitochondria.
https://www.youtube.com/watch?feature=player_embedded&v=KXsxJNXaT7w
Respiration
The cascade of electrons transfers (redox reactions), that culminates in the reduction of the terminal electron acceptor, is called respiration. The amount of energy released by these redox reactions, and thus the amount of energy available for ATP synthesis, depends on the redox potential of the terminal electron acceptor. Oxygen (O2) has the greatest redox potential, and thus aerobic respiration results in the most ATP synthesized. Bacteria and Archaea can use other terminal electron acceptors with lower redox potential when oxygen is not available. This anaerobic respiration produces less ATP.
Bacteria can modify their electron transport chains to use a variety of electron donors and electron acceptors, and will switch to the best available electron sources and sinks available in their environment. In marine sediments, microbial communities stratify according to redox potential. The deeper, more anoxic layers use electron acceptors with progressively lower reducing potential.

Chemiosmosis – this is really important!
All prokaryotic cells (bacteria and archaea) maintain a proton gradient (pH gradient) across their plasma membranes (in mitochondria, the proton gradient is across the inner mitochondrial membrane). The interior of the cell is relatively alkaline, whereas the exterior periplasmic space is relatively acidic. A 1000-fold difference in proton concentrations across the plasma membrane results in a proton motive force, consisting of both the chemical concentration gradient of protons and a voltage gradient across the membrane (the protons are positively charged, so an imbalance of protons means that the side of the membrane with more protons has a positive charge relative to the side with fewer protons). This proton motive force is a form of stored energy, and protons returning across the membrane down their concentration and voltage gradients release free energy that can be captured by the cell to make ATP. The lipid bilayer membrane is almost impermeable to protons. The ATP synthase enzyme complex acts like a proton-powered turbine, and couples the energy released from the downhill flow of protons back across the membrane through the ATP synthase rotor to synthesize ATP from ADP and inorganic phosphate. The process of generating the proton motive force across the membrane to power ATP synthesis is called chemiosmosis. In addition to ATP synthesis, prokaryotic cells can use the proton motive force to supply energy for active transport of molecules across the plasma membrane, and to power the motor complex that rotates the bacterial flagellum.
In eukaryotes, mitochondria maintain a proton gradient across the inner mitochondrial membrane and chloroplasts maintain a proton gradient across the thylakoid membrane. Since both these organelles are thought to have originated as prokaryotic endosymbionts, the mitochondrial inner membrane and the thylakoid membrane correspond to the original plasma membranes of the ancestral endosymbionts. Both mitochondria and chloroplasts use the proton gradient for chemiosmotic ATP synthesis and active transport of metabolites into and out of the organelles.
F1 ATP Synthase – know this, too!
Oxidative phosphorylation is the process of chemiosmotic ATP synthesis, using energy from a membrane proton gradient. The F1 ATP synthase enzyme complex that performs this task is located in the membrane, and is a remarkable rotor-stator molecular machine (Stock et al. 1999).
https://www.youtube.com/watch?feature=player_embedded&v=KU-B7G6anqw
The proton motive force drives protons through a channel in the ATP synthase, and turns the rotor at approx 100 rpm. The turning rotor changes the shape of the cytoplasmic subunits, which bind ADP and inorganic phosphate and bond them together to form ATP. Each 360 degree turn of the rotor results in synthesis of 3 ATP molecules. This method of ATP synthesis is called oxidative phosphorylation, because the proton gradient is generated, and maintained, by redox reactions that actively transport protons across the membrane. Oxidative phosphorylation occurs in all respiring cells, even in the absence of oxygen, as long as other terminal electron acceptors are available to drive anaerobic respiration.
The ATP synthases in mitochondria, chloroplasts, and bacteria are all structurally similar, and highly homologous at the molecular sequence level (Watt et al. 2010). A lesser degree of similarity, and more distant homology, exists with archaeal ATP synthases and vacuolar membrane ATPases that function in active transport of protons across the membrane, using the energy from ATP hydrolysis. Indeed, ATP synthases can work in reverse to hydrolyze ATP and replenish the membrane proton gradient.
An evolutionary perspective
The generation of a proton gradient across a membrane and chemiosmosis are universal to life on earth, and are fundamental ways for cells to make a living. Lane and colleagues speculate that “proton power” may have been the earliest form of energy metabolism, essential to, and pre-dating, the last universal common ancestor, LUCA (Lane, 2009; Lane et al. 2010).
The earliest cells, living in anoxic environments, used various electron acceptors. After cyanobacteria caused the Great Oxygenation Event, bacteria that adapted their electron transport chains to exploit oxygen as the terminal electron acceptor gained higher energy yield and thus a tremendous selective advantage. One line of aerobic bacteria took up an endosymbiotic relationship within a larger host cell, providing ATP in exchange for organic molecules. This symbiosis must have occurred in the ancestor of all eukaryotes, because all existing eukaryotes have mitochondria (Martin and Mentel, 2010). The evidence for the endosymbiont origin of mitochondria can be found in:
- the double membrane of mitochondria, where the inner membrane contains the electron transport chain, just as the plasma membrane of the aerobic endosymbiont
- the DNA of mitochondria, whose circular chromosome and genetic sequences resemble the alpha-proteo bacteria.
- the ribosomes of mitochondria, that resemble prokaryotic ribosomes
Powered by hundreds or even thousands of mitochondria, eukaryotic cells attained larger sizes and evolved true multicellular lifestyles.
My video on this topic, 23min. (includes the 3 short segments excerpted above)
Powerpoint slides used in the video above:
B1510_module3_5_respiration_2011Fall
Put it all together: Microbial fuel cells
References:
Lane, N 2009 Was our oldest ancestor a proton-powered rock? New Scientist 19 October 2009 https://www.newscientist.com/article/mg20427306.200-was-our-oldest-ancestor-a-protonpowered-rock.html?page=1
Lane, N, JF Allen, W Martin 2010 How did LUCA make a living? Chemiosmosis in the origin of life. BioEssays DOI 10.1002/bies.200900131
Stock D, Leslie AGW, Walker JE 1999 Molecular architecture of the rotary motor in ATP synthase. Science 286:1700–1705. Abstract/FREE Full Text
Watt, IN, MG Montgomery, MJ Runswick, AGW Leslie, JE Walker 2010 Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria PNAS 107 : 16823-16827 doi:10.1073/pnas.1012260107


It makes sense that F0 part of ATP synthase resembles flagella because flagella acts like motor and gives rotational movement, but how F1 part resembles helicase protein?
Thank you 🙂
The F1 portion resembles helicase in that both have ATPase activity. Both ATPase activities are associated with rotational motion.
A student emailed me the following question, that I thought would be good to share here:
“I have a question regarding some of the material on the electron transport chain and redox potential. I read that electrons are transferred down the ETC consisting of membrane proteins and molecules with increasing redox potential. Does the redox potential increase due to a successively higher concentration of hydrogen protons in the outer membrane? And what is the significance of this larger redox potential?”
Yes, the components of the ETC have increasing redox potential, and the terminal electron acceptor has the largest redox potential. The significance is that the electron transfer reactions from molecules with lower redox potential to molecules with higher redox potential are all exergonic reactions. This means that they will occur spontaneously (no additional energy required), and also that the energy released in these reactions can be coupled to active transport of protons against the concentration gradient. The redox potential is not affected by the proton gradient across the membrane, but is an intrinsic property of the molecules. It may be useful to think of redox potential as a molecule’s tendency to acquire electrons.
Is oxidative phosphorylation and chemiosmosis same thing?
They are closely related; oxidative phosphorylation specifically refers to the synthesis of ATP, powered by a proton gradient generated via chemiosmosis. Chemiosmosis is the transport of protons across the membrane to generate a proton motive force to power oxidative phosphorylation.