Learning Objectives
- Analyze the evidence for DNA as the genetic material through the findings of classical experiments in biology, starting with bacterial transformation
- Describe the key features of the Watson-Crick model of DNA structure, including double helix, base-pairing, antiparallel orientation of strands, and types of bonds
- Predict outcomes from different modes of DNA replication in the Meselson-Stahl experiment
- Describe the basic machinery and process of DNA replication and predict outcomes if a given component of that machinery were missing or nonfunctional
- Explain how the properties of DNA polymerase and the structure of DNA causes DNA to be replicated differently for the leading and lagging strands in bidirectional DNA synthesis
Today we take it for granted that DNA, present as a single chromosome or one of several chromosomes in the cell, is the genetic material that encodes all the information required to “build” an organism. But DNA as the genetic material was once a very controversial idea. Cells are incredibly complex collections of biological macromolecules that carry out diverse biological reactions, while DNA is an extremely simple molecule comprised of only 4 nucleotide bases. How could something so simple contain all the information required to build a cell? An organism?
Part 1: The classic experimental evidence that DNA encodes genetic information
Frederick Griffith discovered bacteria can acquire genetic material from their environment
Frederick Griffith worked with two strains of Streptococcus pneumoniae. The S strain produced a polysaccharide capsule and formed smooth-looking colonies. The R strain could not produce the polysaccharide capsule and formed rough-looking colonies. The S strain was virulent and killed mice. The R strain was non-virulent when injected into mice. Griffith (1928) reported that mixtures of heat-killed extracts of the S cells plus live R cells killed mice, although neither R nor S when injected alone was virulent. Autopsies of the dead mice showed bacterial cells in the blood. When cultured, these formed smooth colonies. He concluded that something from the extract of heat-killed S cells had transformed some of the R cells into S cells. This work defined the concept of bacterial transformation, the ability of bacteria to acquire genetic material from their environment.

Avery, McLeod and McCarty showed that bacterial transformation is due to DNA
Oswald Avery, Colin McLeod, and Maclyn McCarty painstaking purified the transforming substance identified by Griffith and reported in their 1944 paper that the substance was DNA. Their purification procedure eliminated polysaccharides, and they found that the transforming substance was unaffected by proteases or ribonuclease (enzymes that cleave proteins and RNAs), but was destroyed by deoxyribonuclease, an enzyme that destroys DNA. Thus they concluded that transforming substance had to be DNA.
Here is a video summary of the design and conclusions from these two sets of experiments:
Alfred Hershey and Martha Chase showed that bacteriophage DNA, not protein, infects cells
The bacteriophage T2 is a virus that infects E. coli cells. Bacteriophages, viruses that infect prokaryotic cells, are composed only of a protein capsule that contains a DNA molecule.

Hershey & Chase radioactively labeled either bacteriophage proteins (using radioactive sulfur) or bacteriophage DNA (using radioactive phosphate) and showed that labeled DNA entered infected cells, whereas labeled protein stayed outside. “Blending” refers to the use of a kitchen blender to shear off the attached bacteriophage particles from the infected cells. (This experiment is how bar blenders became standard equipment in microbiology and molecular biology labs.)
Here is a video that summarizes the design and conclusions of the Hershey-Chase experiment:
Part 2: The Watson-Crick model of DNA structure
Watson & Crick constructed a model of DNA structure that fits Franklin’s X-ray diffraction data & Chargaff’s rules
The biochemist Erwin Chargaff analyzed the base composition of DNA from a wide variety of species and found that although the percentages of the four nucleotide bases adenine, guanine, cytosine and thymine (A, G, C and T) varied from species to species, the following proportions were always seen:
A = T; G = C
Watson and Crick used Rosalind Franklin’s X-ray diffraction data on purified DNA to deduce that DNA has a helical structure. They were able to solve the structure of DNA when they realized that the molecule was a duplex consisting of two anti-parallel strands, with the sugar-phosphate backbone on the outside and the bases paired on the inside with hydrogen bonds between A and T or between C and G. The pairing of the bases accounted for Chargaff’s rules that the amount of adenine and thymine were the same, and the amount of guanine and cytosine were the same.

What does antiparellel mean? It refers to the directionality of a strand of DNA (or RNA) and can be seen in the sugar-phosphate backbone. One end of the backbone has a 5′ carbon with a phosphate (PO4) attached; the other end has a 3′ carbon with a hydroxyl (-OH) group attached. We indicate directionality as 5′ –> 3′, said “five prime to three prime.” The two strands of DNA are not only complementary to each other (where one strand has an A, the complementary strand has a T, and where one strand has a C, the complementary strand has a G, and so forth) but run in opposite directions: 5′–>3′ and 3′ <– 5′.

Part 3: The Meselson-Stahl experiment revealed the mechanism of DNA replication
Meselson and Stahl demonstrate that DNA replication cannot be conservative or dispersive but must instead be semi-conservative
The structure of DNA, consisting of two complementary strands, suggests that DNA could replicate by unzipping into two separate strands, and each strand would serve as the template for the synthesis of its complementary partner strand. This is called semi-conservative replication, because each resulting daughter DNA molecule would consist of one original (parental) strand and one newly synthesized strand. Other possible modes would be conservative replication, and dispersive replication.
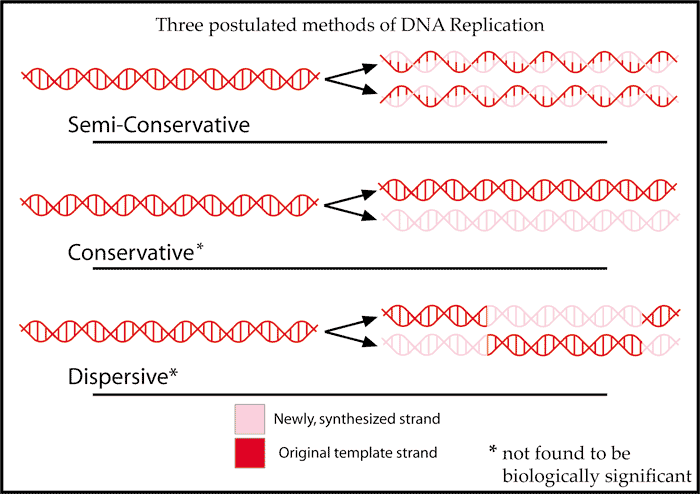
Matthew Meselson and Franklin Stahl used a density isotope of nitrogen, 15N, to label DNA. With density-gradient ultracentrifugation, they analyzed 15N density-labeled DNA (“heavy DNA”) before and after rounds of replication in medium containing normal 14N (“light nitrogen”). The semi-conservative model predicted that, in this experiment, the daughter DNA molecules after one round of DNA replication would all consist of one 15N-labeled DNA strand and one 14N-labeled DNA strand, and therefore would have intermediate density. That was exactly the result observed.

Students should work through the results predicted by conservative and dispersive modes of replication, and explain how the observed results disprove these alternative hypotheses.
Here is a video summary of the Meselson-Stahl experiment:
Part 4: The enzymes of DNA replication and replication differences at the leading vs lagging strand
DNA replication proceeds bidirectionally from an origin of replication, but the two strands are replicated differently
- Replication of chromosomal DNA begins at special sites called origins of replication, where the DNA duplex is unzipped. Origins of replication tend to be full of A-T base pairs rather than G-C. What property of A-T pairing vs G-C pairing could explain this observation?
- DNA replication proceeds bidirectionally from the origin, with the enzyme helicase at the replication “fork” unzipping and unwinding the DNA template in both directions away from the origin.
- DNA polymerase requires an RNA primer, synthesized by primase, to begin polymerizing DNA. DNA polymerase has to have an existing 3′-OH to add to, so it cannot start synthesizing a complementary strand from scratch. But RNA polymerase can start from scratch, so a new DNA strand starts with an RNA primer which is synthesized by primase. DNA polymerase then starts by adding on to the 3′-OH end of the RNA primer.
- DNA polymerase adds new nucleotides only at the 3′-OH end, so new strands grow 5′ –> 3′.
- DNA polymerase reads the template strand 3′ –> 5′, so the new DNA duplex is antiparallel.
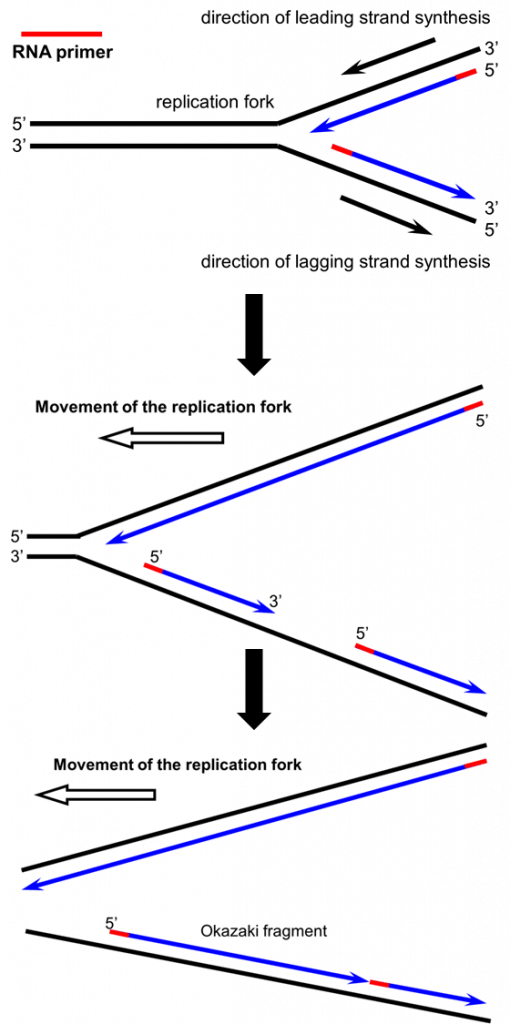
- One daughter strand, called the “leading” strand, can be synthesized continuously, in the same direction that the DNA is being unzipped at the fork.
- The other daughter strand, the “lagging” strand, has to be synthesized in short fragments, called “Okazaki” fragments, that start with an RNA primer and are synthesized going away from the fork.
- The Okazaki fragments are sealed by an enzyme called DNA ligase.

As the replication fork extends in one direction above, the leading strand (top strand) can be synthesized continuously because it is proceeding in the 5′ to 3′ direction. The lagging strand must be synthesized in short fragments as the DNA fork extends. This is because DNA synthesis must ALWAYS proceed in the 5′ to 3′ direction (adding the 5′ phosphate end of a new nucleotide to the 3′ hydroxyl group of the last nucleotide in the existing chain), but the lagging strand template is running the ‘wrong’ way for synthesis to occur in the direction of the replication fork. Note both strands require RNA primers (red) to begin DNA synthesis.
In the panels below, you can see movement of the replication fork “in action,” and why the leading strand has continuous synthesis while the lagging strand is composed of short fragments based on the direction of DNA synthesis in the 5′ to 3′ direction:
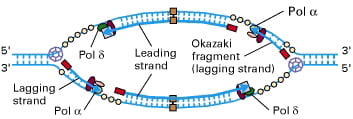
In the image below, you can see that each side of the replication “bubble” or “eye” has two replication forks, and each fork has a leading and a lagging strand:
Summary videos
This video provides an engaging summary of the ideas discussed above for DNA structure and the enzymes involved in DNA replication (see if you can spot the mistake on DNA structure):
For more help with these topics, you can view Dr. Choi’s video lectures below. The experiments proving that DNA is the hereditary material, and the structure of DNA:
The Meselson-Stahl experiment and DNA replication:
Slide set to go with the two lecture videos on this page: B1510_module4-5_DNA_2011
Put it all together: forensic_dna_sequencing_2012
Other resources
Molecular animation of DNA replication:
https://www.dnalc.org/resources/3d/04-mechanism-of-replication-advanced.html
Sustainable Development Goal

UN Sustainable Development Goal (SDG) 9: Industry, Innovation, and Infrastructure – The properties of DNA polymerase and the structure of DNA are important in the development of DNA sequencing technologies and biotechnology products. These technologies have the potential to revolutionize various industries, including healthcare, agriculture, and environmental science.


I believe Hank Green’s mistake was that he circled the 5′ end every time.
Yes, he circles the 5′ end and calls it the 5′ end the first time, then he circles the 5′ carbon on the next nucleotide and calls it the 3′ – about 3:14 into the video.
I have a question:
Is there a reason for having the two DNA strands anti-parallel? Is it advantageous in any way?
I believe the base-pairing (hydrogen bonds) doesn’t work if the two strands are parallel. The two strands have to be antiparallel for the bases to be in the right orientation to form the hydrogen bonds.
‘Writing to his son, Michael, Francis Crick tells of a “most important discovery.”’
http://www.nytimes.com/interactive/2013/02/26/science/crick-letter-on-dna-discovery.html?smid=fb-share&_r=0