Learning objectives
- Diagram and explain the Central Dogma of molecular biology
- Identify the general functions of the three major types of RNA (mRNA, rRNA, tRNA)
- Identify the roles of DNA sequence motifs and proteins required to initiate transcription, and predict outcomes if a given sequence motif or protein were missing or nonfunctional
- Predict the RNA transcribed from a DNA sequence identified as either the template strand or the coding strand, and vice versa
- Use the genetic code to predict the amino acid sequence translated from an mRNA sequence
- Describe the process of and key components required for translation.
- Differentiate between types of DNA mutations, and predict the likely outcomes of these mutations on a protein’s amino acid sequence, structure, and function
- Compare and contrast prokaryotic and eukaryotic transcription and translation
The Central Dogma
Francis Crick coined the phrase “the Central Dogma” to describe the flow of information from nucleic acid to protein. Information encoded in DNA is transcribed to RNA, and RNA is translated to a linear sequence of amino acids in protein. Although information can flow reversibly between DNA and RNA via transcription and reverse transcription, no mechanism has yet been found for alterations in protein amino acid sequence to somehow effect a corresponding change in the RNA or DNA.
The DNA in the central dogma encodes a gene, a chain of nucleotides that is transcribed to produce a functional RNA. In Mendelian genetics, we also call a gene the basic unit of heredity.
This video gives a highly simplified overview of the central dogma of molecular biology:
And this video provides an animated overview of gene expression in a eukaryotic cell:
Transcription: DNA to RNA
Transcription is the process of using DNA as a template to make an RNA molecule:
- The enzyme RNA polymerase reads the template strand of DNA and synthesizes an RNA molecule whose bases are complementary to the template strand of DNA.
- RNA is synthesized 5′ –> 3′ (same direction as DNA synthesis); RNA polymerase reads the template strand of DNA 3′ –> 5′.
- The sequence of bases in RNA is the same as the sequence of bases in the “coding” strand of DNA, except that RNA has uracil (U) instead of thymine (T).
- RNA polymerases in both prokaryotes and eukaryotes depend on DNA-binding proteins, called transcription factors, to bind to special sequence motifs in the DNA called promoters, to recognize where genes start. A DNA sequence motif is a short, recurring nucleotide pattern that has (or is presumed to have) a biological function.
- Transcription factors recruit RNA polymerase to bind to the promoter sequence and begin transcription just “downstream” of the promoter.
This video gives a simplified overview of transcription. Notice that the narrator makes a mistake at 3:45 (that he later catches and corrects!); this mistake serves as a really important reminder of one of the major differences between DNA and RNA, so watch for it.
See a more advanced molecular animation of transcription, with narration, here: https://www.dnalc.org/resources/3d/13-transcription-advanced.html
Translation: RNA to Protein
Translation is the process of using an mRNA molecule as a template to make a protein:
Translating a sequence of bases in the RNA to a sequence of amino acids in proteins requires 3 major components:
- messenger RNA (mRNA): mRNAs are transcribed from protein-coding genes to make an RNA copy of the protein-coding parts of the gene. (There are other types of genes which do not encode proteins, such as genes encoding rRNAs and tRNAs.)
- ribosomes: ribosomes are large assemblies of ribosomal RNA molecules (rRNAs) and dozens of proteins that function in gene translation to assemble the amino acid chain by reading the code in the mRNA. When they are not working, they fall apart into the small subunit and large subunit, each consisting of a rRNA and numerous proteins. When the structures of prokaryotic ribosomes were determined at high resolution, researchers were astonished to discover that the catalytic site for the peptidyl-transfer reaction (attaching new amino acids to the growing polypeptide chain) consists entirely of rRNA. Thus the ribosome is actually an immense ribozyme, or a catalytic RNA molecule stabilized by numerous proteins, rather than an enzyme.
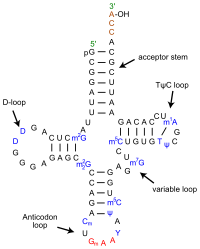
- transfer RNAs (tRNAs): tRNAs bring the amino acids to the ribosome during protein translation. Each tRNA is “charged” with its corresponding amino acid dictated by the anticodon loop of the tRNA. A charged tRNAs is attached to its corresponding amino acid, while an uncharged tRNA is not carrying an amino acid. tRNAs match the amino acid to the codon in the mRNA. The bases in the anticodon loop are complementary to the bases in an mRNA codon. The 3′ end of the tRNA has a high-energy bond to the appropriate amino acid. Cells have a family of enzymes, called amino-acyl tRNA synthetases, that recognize the various tRNAs and “charge” them by attaching the correct amino acid.

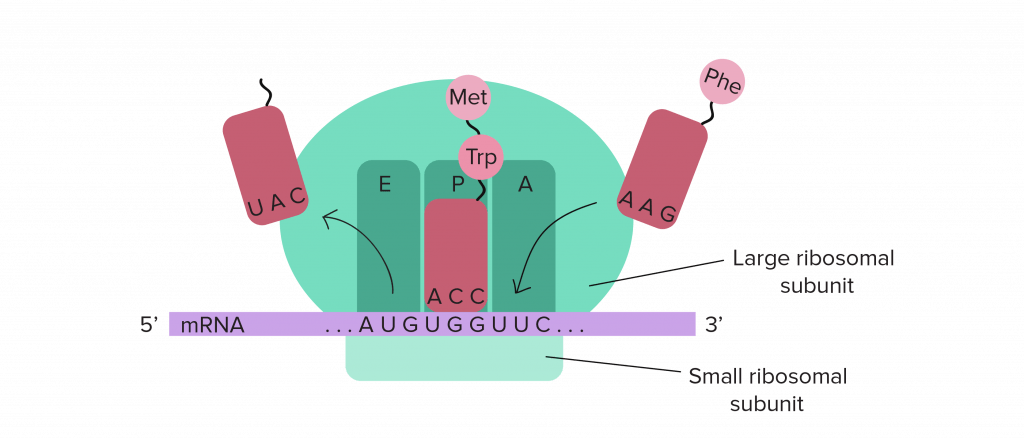
Translation begins near the 5′ end of the mRNA, with the ribosomal small subunit and a special initiator tRNA carrying the amino acid methionine. In most cases, translation begins at the AUG triplet closest to the 5′ end of the mRNA. In eukaryotes, the small subunit of the ribosome typically just “scans” along from the 5′ end of the mRNA until it finds the first AUG codon. In prokaryotes, there is typically a specific sequence that the ribosome binds to, which “positions” the ribosome at the starting AUG. In either case, the large ribosomal subunit then docks and translation begins, always starting with an AUG codon (methionine) in both prokaryotes and eukaryotes. The ribosome moves along the mRNA 3 bases at a time, from the 5′ to the 3′ direction, and new tRNAs whose anti-codons are complementary to the mRNA codons arrive with their corresponding amino acids. A peptide bond forms to join the amino acid to the carboxyl end of the growing polypeptide chain. The ribosome moves another 3 bases, and the empty tRNA is ejected to make room for a new amino-acyl tRNA.
As with DNA, the polypeptide chain made by the ribosome also has directionality; one end has a free amino group and the other end of the chain has a free carboxyl group. These are called the N-terminus and the C-terminus, respectively. New amino acids are added only to a free carboxyl end, so polypeptide chains grow from the N-terminus to the C-terminus.
This video gives a solid overview of translation. It is a little longer than the typical videos we use in the book, but it does a really nice job of breaking down step-by-step what happens during translation:
Watch a much shorter molecular animation of translation here:
https://www.dnalc.org/resources/3d/16-translation-advanced.html
The Genetic Code
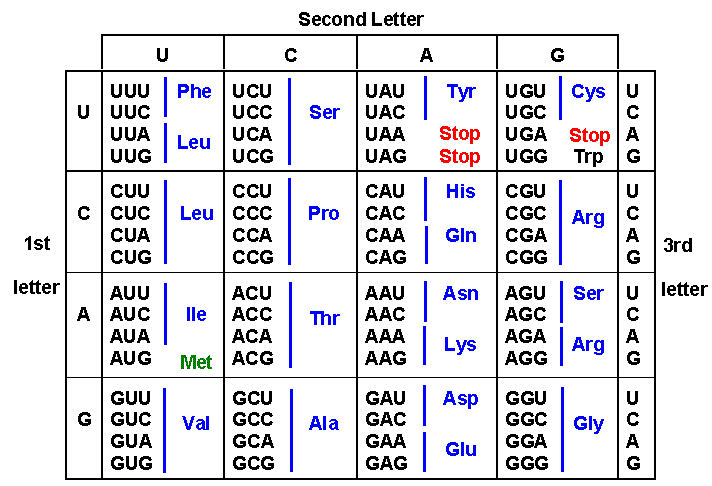
This genetic code is universally used by all living organisms, whether Archaea, Bacteria or Eukarya, with only minor modifications in the mitochondria of a relatively few species. If you do the math, you can see that there are 64 possible codons (4^3), but we know that there are only 20 amino acids. Thus the code is “degenerate,” because the same amino acid can be specified by 2, 3, 4 or even 6 different codons. For example, glycine can be specified by codons GGU, GGC, GGA, and GGG. Methionine is unusual in that it is specified only by a single codon: AUG. (Tryptophan is the only other amino acid specified by a single codon.)
Mutations can have vastly different effects depending on where they occur in a gene, and where they occur in a codon
If we consider just single nucleotide changes (substitutions, deletions, or insertions of single bases), these can have very different consequences depending on where they occur in the gene. Often a DNA base substitution will have no effect if they change the 3rd base in the codon, because the 3rd position for most amino acids is redundant, given the degenerate nature of the genetic code. For example, changing GAG to GAA has no effect on the protein because both codons specify alanine. Such “silent” mutations are called “synonymous” mutations.
Other base substitutions in the 1st or 2nd position will cause amino acid changes; these are “nonsynonymous” mutations, also called mis-sense mutations. Even among nonsynonymous mutations, the exact amino acid change matters. A change of one hydrophobic amino acid to another hydrophobic amino acid will be less disruptive to the structure of the protein than a change of a hydrophobic amino acid to a polar or charged amino acid. Finally, some parts of a protein are more important than others, such as the catalytic site of enzymes, or sites that bind other proteins, DNA, or regulatory molecules.
Some mutations create a new stop codon (UAA, UAG, or UGA). These are called “nonsense” mutations and cause truncated polypeptides to be made. Insertions or deletions (“indels”) of single nucleotides cause a change in the reading of all downstream codons; they are shifted by one base. Such “frameshift” mutations will alter most or all amino acids downstream (towards the 3′ end of the mRNA, towards the C-terminus of the protein) of the mutation. Both stop codon mutations and frameshift mutations will have predicted large effects on protein function if they happen early in the sequence.
Differences between prokaryotes and eukaryotes
Much of what is discussed above was originally discovered in bacteria and later found to be true of archaea and eukaryotes as well; many of the core features of molecular biology are evolutionarily conserved. However, there are a few key differences as outlined below.
Prokaryotes: transcription and translation are coupled
In prokaryotic cells, ribosomes begin to translate even while the mRNA is still being transcribed. DNA, RNA polymerase, and ribosomes are all in the same location. This coupled transcription and translation can occur because prokaryotes have no nucleus. In eukaryotes, the nucleus separates the transcription machinery from the translation machinery.
Eukaryotes: transcription and translation are separated in space and time, and nuclear pre-mRNA undergoes processing to become mature mRNA
In eukaryotes transcription occurs in the nucleus, whereas translation occurs outside the nucleus in the cytoplasm by free cytoplasmic ribosomes or by ribosomes docked to the ER. The RNA transcribed from a protein-coding gene in the nucleus is called the pre-mRNA. Pre-mRNA has to undergo at least two, and usually 3, processing steps before they can be exported to the cytoplasm as mature mRNA. These are, in order:
- The 5′ end of the pre-mRNA is modified by the covalent attachment of a 7-methylG nucleotide, called the 5′-cap. The 5′ cap is required for eukaryotic ribosomes to initiate translation.
- The majority of eukaryotic genes contain sequences which do not actually code for protein. These sequences are called introns (“intervening” sequences), and they “interrupt” the protein coding sequences, which are called exons (“expressed” sequences) in the gene. These non-protein coding intron sequences are removed by RNA splicing, leaving just the protein-coding exons in the final mRNA.
- The 3- end of the pre-mRNA is modified by the addition of hundreds of adenine nucleotides, called the polyA tail. The polyA tail is important for nuclear export, mRNA stability, and translation.
All of these processing steps actually happen while the mRNA is being transcribed. The techincal term is that these steps occur co-transcriptionally. Because of this, a full-length “pre-mRNA” never actually exists.
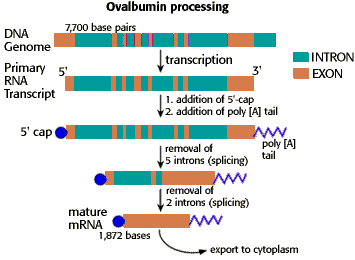
This video gives a nice quick overview of these differences between prokaryotes and eukaryotes:
Resources
Here is Dr. Choi’s video lecture on this topic:
Test your knowledge with these questions & problems: DNA_to_protein_questions
And the slide set: B1510_module4-6_DNA_to_protein
Sustainable Development Goal

UN Sustainable Development Goal (SDG) 7: Affordable and Clean Energy – Knowledge of the DNA sequence motifs and proteins required to initiate transcription can lead to the development of biotechnology that can produce biofuels and other forms of clean energy. Frequently, biofuel advances are made through inserting new genes to be expressed, or increasing the expression of current genes, in the plants and bacteria used to maximize yields.



So DNA template strands have promoters which is where the RNA polymerase binds in order to start transcription, but what is it that stops transcription?
There are also transcription termination signals in the DNA.