Learning Objectives
- Describe the role of protein:DNA interactions in regulating transcription initiation in prokaryotes and eukaryotes
- Distinguish positive regulation from negative regulation
- Use models of gene regulatory systems such as the lac operon to predict the effects of mutations in various components
- List post-transcriptional modes of regulation in eukaryotes
Cells express (transcribe and translate) only a subset of their genes. Cells respond and adapt to environmental signals by turning on or off expression of appropriate genes. In multicellular organisms, cells in different tissues and organs become specialized by making different sets of proteins, even though all cells in the body (with a couple of exceptions) have the same genome. Such changes in gene expression, or differential gene expression among cells, are most often regulated at the level of transcription.
Promoters and upstream regulatory sequences
Promoters are sites in the DNA where RNA polymerase binds to initiate transcription. Promoters also contain, or have near them, binding sites for transcription factors: DNA-binding proteins that can either help recruit, or repel, RNA polymerase. The promoter along with nearby transcription factor binding sites constitute an upstream regulatory sequence or region.
https://www.youtube.com/watch?v=MkUgkDLp2iE
E. coli lac operon: dual positive and negative regulation

The lac operon of E. coli has 3 structural genes:
- lacZ encodes the enzyme beta-galactosidase, which cleaves lactose into glucose and galactose
- lacY encodes permease, a membrane protein for facilitated diffusion of lactose into the cell
- lacA encodes transacetylase, and enzyme that modifies lactose
An mRNA encoding all 3 proteins is transcribed at high levels only when lactose is present, and glucose is absent.
Negative regulation by the Repressor – In the absence of lactose, the lac Repressor protein, encoded by the lacI gene from a separate promoter that is always active, binds to the Operator sequence, located between the promoter and the transcription start site. Repressor bound to the Operator presents RNA polymerase from initiating transcription.
When lactose is present, an inducer molecule derived from lactose binds allosterically to the Repressor, and causes the Repressor to leave the Operator. RNA polymerase is then free to initiate transcription.
Positive regulation by CAP – Glucose is the preferred substrate for energy metabolism. When glucose is present, cells transcribe the lac operon only at very low levels, so the cells obtain most of their energy from glucose metabolism. RNA polymerase by itself binds rather poorly to the lac promoter.
Glucose starvation causes a rise in the level of cyclic adenosine monophosphate (cAMP), an intracellular alarm signal. cAMP binds to the catabolite activator protein (CAP). The CAP-cAMP complex binds to a site near the lac promoter and recruits RNA polymerase to the promoter.
High level transcription of the lac operon requires that CAP-cAMP be bound to the CAP binding site, and that Repressor is absent from the Operator. These conditions normally occur only in the absence of glucose and presence of lactose.
Positive regulation and negative regulation are universal themes for gene regulation in both prokaryotes and eukaryotes.
Complexities of eukaryotic gene regulation
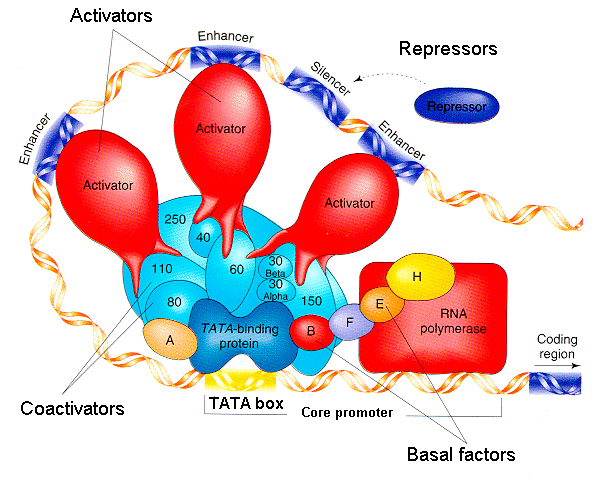
Eukaryotic gene regulation is generally much more complex than prokaryotic gene regulation. The upstream regulatory regions of eukaryotic genes have binding sites for multiple transcription factors, both positive regulators and negative regulators, that work in combination to determine the level of transcription. Some transcription factor binding sites, called enhancers, work at quite a distance, tens or hundreds of thousands of base pairs away from the promoter.
As a general rule, eukaryotic genomes do not have genes arranged in operons. Instead, eukaryotic genes that are co-regulated tend to have the same DNA regulatory element sequence associated with each gene, even if those genes are located on completely different chromosomes. This means that the same transcriptional activator or repressor can regulate the transcription for every single gene that has that particular DNA regulatory element associated with it.
Another major difference between prokaryotic gene regulation and eukaryotic gene regulation is that the eukaryotic (but not prokaryotic) DNA double helix is organized around proteins called histones which organize the DNA into nucleosomes. This combination of DNA + histones is called chromatin.
Chromatin can be condensed in a 30-nm fiber formation (tightly compacted nucleosomes) or loosely arranged as “beads-on-a-string,” where the DNA between and around nucleosomes is more accessible. This compaction is controlled by post-translational modifications which are added to the histones in the nucleosomes. When histones have acetyl groups added to them by enzymes called histone acetyl transferases (HATs), the acetyl groups physically obstruct the nucleosomes from packing too densely and help to recruit other enzymes that further open the chromatin structure. Conversely, when the acetyl groups are removed by histone deacetylases (HDACs), the chromatin assumes a condensed formation that is transcriptionally inactive. Transcription factors work in combination to recruit and direct histone modifying enzymes (HATs and HDACs) to enable or disable transcription of their target genes.
In addition to regulating transcription, eukaryotic cells can regulate gene expression at any step after transcription, including RNA splicing, export from the nucleus, translation, and post-translational modification of proteins.
Overall differences and similarities
If you understand the similarities and differences in eukaryotic and prokaryotic gene regulation, then you know which of the following process are exclusive to eukaryotes, which are exclusive to prokaryotes, which occur in both, and how each is accomplished:
- coupled transcription and translation
- 5′ cap and 3′ poly(A) tail
- AUG as the translation initiation codon
- regulation of gene expression by proteins binding to DNA regulatory elements
- alternative mRNA splicing
- regulation of gene expression through chromatin accessibility