Learning Objectives
- Distinguish organic molecules from inorganic molecules
- Identify the 4 major molecular components of biomass
- Match each biological macromolecule with the type of subunit building block, and the bonds that link the subunits into polymers
- Identify the main cellular functions for each type of macromolecule
- Distinguish between DNA and RNA
- Identify the 4 levels of structure in proteins, and what bonds, forces or interactions are responsible for each level of structure (primary, secondary, tertiary, quaternary)
- Differentiate among properties of different types of amino acid R-groups and relate how an amino acid change would affect the structure and function of a protein
Before we begin, we assume that you know these basic chemical concepts:
- Atoms and elements
- Periodic table of the elements
- Valence electrons
- Electronegativity
- Chemical bonds
If you are unfamiliar with them, you should review our web page: Chemical context for Biology. You can also read a fuller explanation in the free OpenStax Biology textbook: 2.1 Atoms, Isotopes, Ions and Molecules: the Building Blocks
1. All living organisms are made of organic molecules.
One of the distinguishing features of life is that cells are made of organic compounds and large molecules constructed from simple organic compounds. Up to the early 19th century, scientists thought only living organisms could make organic compounds. Organic compounds are all built from carbon atoms, but not all molecules containing carbon are organic. So how do we recognize organic molecules?
- Organic molecules must have C and H, and may have O, N, P, S (a handy mnemonic is CHNOPS for carbon, hydrogen, nitrogen, oxygen, phosphorus and sulfur)
- Organic molecules have at least one covalent bond between C and H or between C and C. In chemistry parlance, the carbon atom in organic molecules must be reduced and not be fully oxidized. A fully oxidized carbon is not organic because it would have covalent bonds only to oxygen atoms. For instance, carbon dioxide (CO2; O=C=O) is an inorganic form of carbon because the carbon atom has bonds only to oxygen atoms, and is therefore completely oxidized.
- (An exception to the above rules is urea, where carbon has bonds to 2 amino nitrogens and a double bond with oxygen – but we won’t ask you to remember this exception; focus on the general rules above.)
Organic molecules can arise naturally from abiotic synthesis (recall the Miller-Urey experiment), but in the biosphere, most organic molecules are synthesized by living organisms.
Synthesis of organic carbon molecules from inorganic CO2 requires energy and chemical reducing power, as the carbon atoms in organic molecules are in reduced form. For a review of oxidation-reduction (redox) reactions from a biological perspective, see this Khan Academy video. Briefly, atoms such as carbon or oxygen are said to be reduced if they form covalent bonds with an atom with lesser electronegativity, such as hydrogen. Conversely, carbon is oxidized when it forms a covalent bond with an atom with greater electronegativity, such as oxygen. Recall that a covalent bond is formed when two atoms share a pair of electrons. The reduced atom has gained a majority share of the electrons that form the covalent bond, and the oxidized atom has only a minority share.
2. The biomass of a cell (the organic contents, excluding water and inorganic salts) is composed of 3 types of macromolecules, plus lipids.
The 3 types of macromolecules (very large molecules) are polysaccharides, nucleic acids (DNA and RNA), and proteins. You should know how cells make these macromolecules, and their basic structures and functions.
3. Small organic molecules are covalently linked (polymerized) to form the 3 types of large biological macromolecules (polymers); lipid membranes self-assemble.
One recent study concluded that cells are composed of 68 distinct organic molecules (Marth 2008) that are assembled into 3 biological polymers plus lipid structures (membranes). Polymerization of monomers into polymers occurs by dehydration reactions, chemical reactions that link two subunits together via a covalent bond while extracting an -OH and an H to create a molecule of water: H2O. So dehydration reactions remove a molecule of water from the starting molecules in the process of forming a covalent bond between those starting molecules.
Cleavage of polymers back to monomers occurs by hydrolysis reactions, where a molecule of water is split (hydrolyzed) to -OH and H. Hydrolysis reactions break the bonds linking two subunits. This is exactly the reverse of a dehydration reaction. See the diagrams below on glycosidic bonds and peptide bonds to see how water molecules are created or used in these reactions.
Lipids, by definition, are water-insoluble organic molecules. Lipids in water can spontaneously aggregate via hydrophobic interactions to form lipid bilayer membranes. Hydrophobic interactions arise from nonpolar molecules avoiding water—having all the nonpolar molecules associate together minimizes their interaction with water.
How can we predict whether an organic molecule will be hydrophobic (a lipid) or hydrophilic? If the molecule has negatively or positively charged atoms (is ionized), or has a high proportion of polar bonds (C-O or C-N), then the molecule is hydrophilic. If the molecules has mostly non-polar bonds (C-H or C-C), then it is hydrophobic.
Below are descriptions of the 3 types of macromolecules and lipid membranes:
a) Polysaccharides are polymers made by linking monosaccharides via glycosidic bonds (see figure below). Examples are starch, cellulose, and chitin. Monosaccharides are organic molecules with the composition [CH2O]n, where n is usually 3-6. For instance, glucose is a 6-carbon sugar with the formula C6H12O6. Other examples include 5-carbon sugars like ribose. The term carbohydrates may refer either to monosaccharides of the composition [CH2O]n or to polysaccharides. Complex carbohydrates often have branched structures.
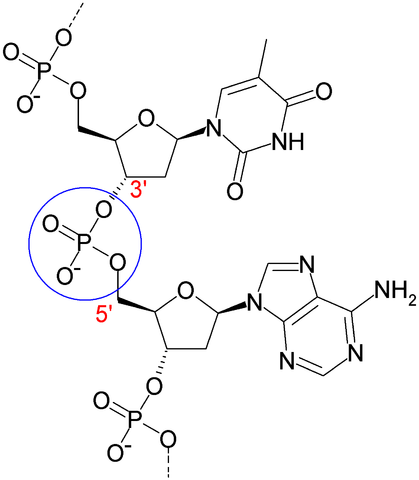
b) Nucleic acids (RNA and DNA) are polymers made by joining nucleotides (5-carbon sugar-phosphate + nitrogenous base) in a phosphodiester linkage. Hydrogen bonding between paired bases (A:T and G:C) stabilize DNA duplexes and RNA secondary structures that form by intra-molecular base pairing (A:U and G:C).
c) Polypeptides (proteins) are polymers of amino acids, joined together by peptide bonds. Peptide bonds are formed between the carboxyl group (carbon with 2 oxygen atoms bonded to it) of one amino acid and the amino group (nitrogen with 2 hydrogen atoms) of the next amino acid. All amino acids have a nitrogen, an alpha-carbon with a side chain (the R group in the diagram below – the 20 different amino acids differ in their R groups), and a carboxyl carbon. The nitrogen, alpha-carbon and carboxyl carbons form the peptide backbone of a polypeptide chain. Figures of protein structures often show only the peptide backbone, leaving out the side-chain R groups.
How proteins fold into their overall 3-dimensional structures, and interact with each other to form larger multi-protein complexes, are determined by various bonds and interactions, as described below (section #6).
Self-review: You should be able to distinguish among these macromolecules and identify the monomers that build each type of macromolecule.
d) Living organisms also contain lipid bilayer membranes made of phospholipids. The phospholipids spontaneously self-assemble in water to form bilayer membranes, via hydrophobic interactions.
The phospholipid bilayers create boundaries and a hydrophobic environment that separates the internal aqueous environment of the cytosol from the outside of the cell, and also separates distinct intracellular organelle compartments in eukaryotic cells. Membranes make it possible for cells to create and maintain large differences in ion concentrations that drive cellular energy metabolism, to regulate transport of materials and water into and out of the cell, and to receive and sense extracellular signals.
4. Cells use the different classes of biological macromolecules in different ways.
a) Polysaccharides are used primarily for energy storage (glycogen, starch) and static structures (such as cellulose, chitin), but can also play important roles in cell-cell recognition/adhesion and signaling.
b) Proteins are used primarily for enzymatic activities, signaling, and dynamic structural components.
c) Nucleic acids are used for genetic information storage (DNA or RNA) and retrieval (mRNA). Some RNAs play key catalytic roles in information processing (RNA splicing, protein synthesis).
d) Lipids are used to define the cell’s boundary, compartmentalize the cell (in eukaryotes), for energy storage (triglycerides: fats and oils), and signaling (steroid and other lipid hormones).
5. Cells have two types of nucleic acids: DNA and RNA, that differ in key ways
DNA has bases A, C, G, and T, deoxyribose, and two strands that form a duplex via hydrogen bonds between the bases on one strand and the complementary bases on the partner strand. The primary function of DNA is storage and transmission of hereditary information.
RNA has bases A, C, G and U, ribose, and one strand that may form internal duplexes (called RNA secondary structure) by folding upon itself. In cells, RNA functions in expression of genetic information in DNA to make proteins (mRNA, tRNAs, rRNAs, and other small RNA molecules), but may also serve for storage of hereditary information in many viruses (e.g., influenza, HIV, Ebola).
Study tip: Make a table to highlight the differences between DNA and RNA.
Summary table
| Major biomass components | Subunits | Primary elemental composition* | Major Functions |
| Lipids | hydrocarbons | C, H | membranes energy storage signaling |
| Carbohydrates | monosaccharides | C, H, O | energy storage static structures cell adhesion |
| Proteins | amino acids | C, H, O, N, S | enzymes dynamic structures signaling |
| Nucleic acids | nucleotides | C, H, O, N, P | hereditary information storage and processing |
*Any of these molecules may have modifications or be linked to other molecules that include O, N, P, or S – the elemental compositions are given for the basal molecule types.
6. Protein structures can be described at 4 levels
Among all the biological macromolecules, proteins have the most complex and dynamic structures. Many proteins consist of just a single polypeptide chain. Many other proteins consist of two or more polypeptide chains that must assemble properly to form a functional complex. The function of a protein is determined by its structure; a change in the protein’s activity involves a change in some aspect of the protein’s structure. What, then, determines a protein’s structure?
Each polypeptide is assembled as a linear chain of amino acids covalently linked by peptide bonds. As this chain is being assembled (each subsequent amino acid is bonded onto the free carboxyl-terminus of the nascent polypeptide chain), the polypeptide chain begins to fold.
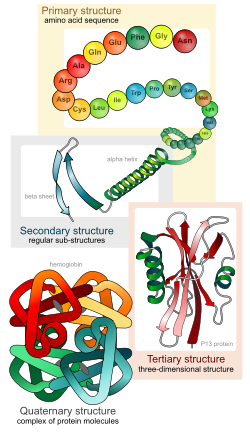
Biologists distinguish 4 levels of protein structure. You should be able to identify the four levels of protein structure, and the molecular forces or interactions responsible for stabilizing each level of structure.
Four levels of protein structure
- Primary structure – the linear sequence of amino acids, held together by covalent peptide bonds.
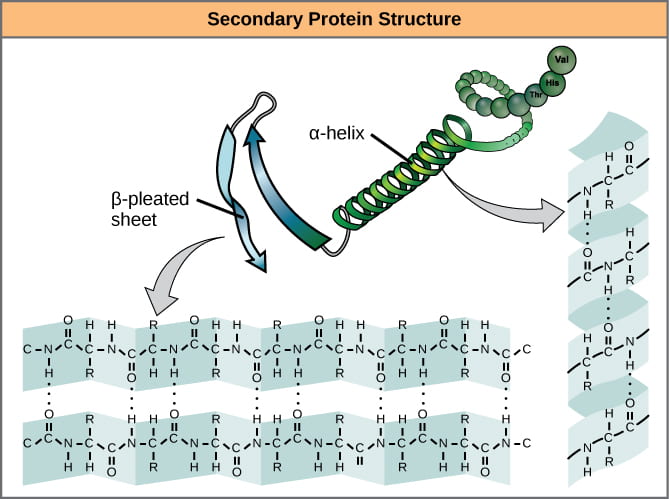
- Secondary structure – alpha helices and beta sheets, stabilized by hydrogen bonds between peptide backbone amino groups and carboxyl groups of amino acids within the same polypeptide chain, but not immediately next to each other. Note that the side-chain R groups are not involved in bonds that stabilize secondary structures.
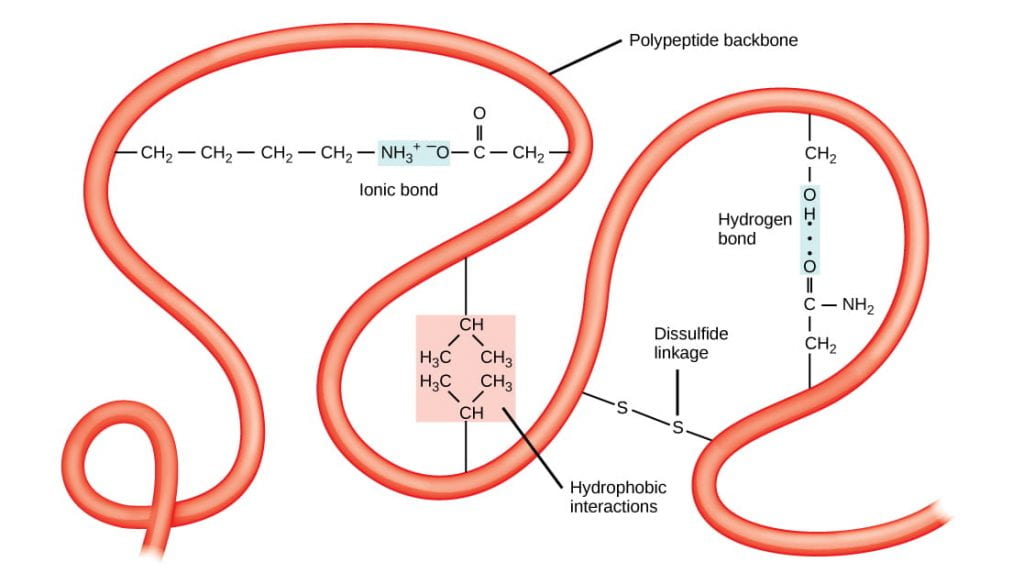
- Tertiary structure – overall 3-D shape of the folded polypeptide chain, that can be described as the spatial relationships of the secondary structure elements linked by loops. Stabilized by various types of amino acid side chain (R-group) interactions, including: hydrophobic and van der Waals interactions, hydrogen bonds, ionic bonds, covalent disulfide bonds between cysteine residues, and interactions with solvent water molecules.
Quaternary structure – assemblage of two or more folded polypeptides into a functional protein unit. Stabilized by inter-chain hydrophobic and van der Waals interactions, hydrogen bonds, ionic bonds, and covalent disulfide bonds between cysteine residues on different polypeptide chains. Proteins that consist of a single polypeptide chain do not have quaternary structure; only proteins that have two or more polypeptide chains have quaternary structure.
7. Changes in the amino acid sequence (primary structure) of a protein can change the structure of the protein and the way it functions.
Case example 1: Hemoglobin:
The classic case exploring protein structure is hemoglobin. Functional hemoglobin is a tetramer, consisting of two alpha-globin and two beta-globin polypeptide chains. Hemoglobin also requires a cofactor, heme, containing an iron atom that binds oxygen.
Questions to be answered after watching the video above:
- What levels of protein structure does hemoglobin exhibit?
- The most common sickle-cell disease mutation changes a glutamic acid (a negatively charged amino acid) in beta-globin to valine (a hydrophobic amino acid). Where would you most commonly expect to find a charged amino acid like glutamic acid, in the interior of the folded protein, or on the surface, interacting with solvent water molecules?
- Which of the following changes do you think might also cause sickle-cell disease?
- the glutamic acid changes to an aspartic acid, a different negatively charged amino acid
- the glutamic acid changes to a lysine, a positively charged amino acid
- the glutamic acid changes to a tryptophan, a hydrophobic amino acid
- the glutamic acid changes to a serine, an uncharged, hydrophilic amino acid
- Sickle cell hemoglobin mutations alter what levels of protein structure (when sickling of red blood cells is apparent)?
Case example 2: Cystic fibrosis
The most common mutation associated with cystic fibrosis causes a single amino acid, a phenylalanine, to be omitted from the protein called CFTR (cystic fibrosis transmembrane conductance regulator). The CFTR protein functions as a chloride channel in the membrane, formed as the single long CFTR polypeptide chain crosses the membrane back and forth several times. The absence of this phenylalanine, which has a large hydrophobic side chain, causes the protein to be mis-folded. Most of the mis-folded protein is recognized by the cellular quality control system and sent to the cellular recycling center (the proteasome); only about 1 percent of the mis-folded CFTR protein makes it to the proper destination, the plasma membrane. My case study is published as a blog post:
Cystic Fibrosis: A Case Study for Membranes and Transport
Case example 3: Extremophiles
Microbes that live in extreme environments of temperature, salt and pH have proteins that are adapted for structural stability in these extreme environments.
Questions for review, further research and thought:
- Do all living organisms synthesize organic molecules from inorganic molecules?
- What processes created organic molecules before life arose? In what environments?
- Why do proteins have enzymatic activities, but generally polysaccharides and nucleic acids do not?
- Carbon atoms are in their most reduced form in which type of organic molecules: carbohydrates, lipids, proteins or nucleic acids?
- Which macromolecules often have branching structures?
- If you heat a cell extract to near boiling for a few minutes, what will be the effect on the 3 types of biological polymers (polysaccharides, proteins, and nucleic acids)? Think in terms of the bonds responsible for the structures of these molecules. This is what happens when cooking food!
- Expanding from the question above, how will changes in pH or salt concentrations affect solutions of each type of macromolecule? Consider that people preserved food in vinegar and salt before refrigeration became available.
The Powerpoint slides for the videos on this page are in this file: B1510_module3_1a_biomolecules
Works Cited
Marth, J. D. 2008. A unified vision of the building blocks of life. Nature Cell Biology 10:1015. https://doi.org/10.1038/ncb0908-1015
Sustainable Development Goal

UN Sustainable Development Goal (SDG) 9: Industry, Innovation, and Infrastructure – Identifying the structure of proteins, including what bonds, forces, or interactions are involved, can be helpful to understanding under what conditions proteins will not function normally. Knowledge of normal structural elements of natural proteins, as well as how proteins may interact, can inform the development of new drugs and materials used in targeted biotechnology therapies and in the creation of sustainable biofuels.