Learning Objectives
- Explain how we came to know that DNA is the molecule that encodes hereditary information
- Describe the key features of the Watson-Crick model of DNA structure
- Predict outcomes from different modes of DNA replication in the Meselson-Stahl experiment
- Explain how the property of DNA polymerase and the structure of DNA causes DNA to be replicated differently for the leading and lagging strands
Fredrick Griffiths discovers bacterial transformation: bacteria can acquire genetic material from their environment
Working with strains of Streptococcus pneumoniae that infect mice, he found that he could transform a non-virulent strain that forms rough colonies (R cells) into a virulent strain that forms smooth colonies (S cells), when he mixed live R cells with heat-killed extracts from S cells.
Avery, McLeod and McCarty show that DNA, but not protein or polysaccharides, can transform bacteria
Their experiments involved purification and enzymatic degradation to show that the substance responsible for Fredrick Griffith’s bacterial transformation was DNA.
Alfred Hershey and Martha Chase showed that bacteriophage DNA infects cells, not protein
T2 is a virus that infects E. coli cells; viruses that infect prokaryotic cells are called bacteriophage or phage. Bacteriphages are composed of just proteins and a DNA molecule packaged inside the phage. Here is a video of bacteriophage T4 landing on the surface of an E. coli cell and injecting it’s genetic material into the cell:
https://www.youtube.com/watch?v=kdz9VGH8dwY

Left: bacteriophage proteins are labeled with 35S; right: phage DNA is labeled with 32P. Labeled phage were incubated briefly with cells to allow attachment and injection of genetic material into the cells. A bar blender was used to shear off the attached phage shells from the cells. Centrifugation pelleted the cells, leaving the phage shells suspended in the supernatant fraction. Radioactive phosphorus pelleted with the cells, but radioactive sulfur remained in the supernatant. Image from wikipedia, by Thomasione, with GFDL or CC-BY-SA license.
Hershey & Chase radioactively labeled either bacteriophage proteins (using radioactive sulfur) or bacteriophage DNA (using radioactive phosphorus), and showed that labeled DNA entered infected cells, whereas labeled protein stayed outside. (Why are these two radioactive elements specific to protein or DNA?) “Blending” refers to the use of a bar blender to physically break the attached bacteriophage particles from the infected cells. This experiment is the reason kitchen blenders became standard equipment in microbiology and molecular biology labs.
Watson & Crick constructed a model of DNA structure that fits Franklin’s X-ray diffraction data & Chargaff’s rules
The biochemist Erwin Chargaff analyzed the base composition of DNA from a wide variety of species, and found that although the percentages of A, G, C and T varied from species to species, the following proportions were always seen:
A = T; G = C
Watson and Crick used Rosalind Franklin’s X-ray diffraction data on purified DNA to deduce that DNA has a helical structure. They were able to solve the structure of DNA when they realized that the molecule was a duplex consisting of two anti-parallel strands (parallel but running in opposite directions), where the sugar-phosphate backbone was on the outside, and the bases paired on the inside with hydrogen bonds between A and T, or between C and G, accounting for Chargaff’s rules.
The directionality of a strand of DNA (or RNA) can be seen in the sugar-phosphate backbone. One end has a 5′ carbon with a phosphate attached; the other end has a 3′ carbon with a hydroxyl (-OH) group attached. We indicate directionality as 5′ –> 3′. The two strands of DNA are not only complementary to each other (where one strand has an A, the complementary strand has a T and where one strand has a C, the complementary strand has a G, and so forth), but run in opposite directions: 5′–>3′ and 3′ <– 5′.
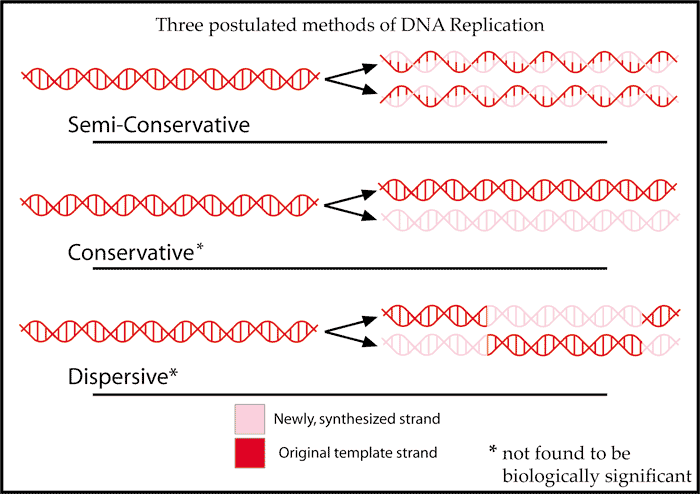
Meselson and Stahl demonstrate that DNA replication cannot be conservative or dispersive, must be semi-conservative
The structure of DNA, consisting of two complementary strands, suggests that DNA could replicate by unzipping into two separate strands, and each strand would serve as the template for the synthesis of its complementary partner strand. This is called semi-conservative replication, because each resulting daughter DNA molecule would consist of one original (parental) strand and one newly synthesized strand. Other possible modes would be conservative replication (the original DNA double helix is reconstituted and the new DNA double helix is 100% new), and dispersive replication (the helix is broken up into pieces during replication and the pieces are attached back together randomly.
Matthew Meselson and Franklin Stahl used a density isotope of nitrogen, 15N, to label DNA and density-gradient ultracentrifugation to analyze 15N density-labeled DNA (heavy DNA) before and after rounds of replication in medium contain normal 14N (light nitrogen). The semi-conservative model predicted that, in this experiment, the daughter DNA molecules after one round of DNA replication would all consist of one 15N-labeled DNA strand and one 14N-labeled DNA strand, and therefore would have intermediate density. That was exactly the result observed.
Work through the results predicted by conservative and dispersive modes of replication, and make sure you can explain how the observed results disprove these alternative hypotheses.
DNA replication proceeds bidirectionally from an origin of replication, but the two strands are replicated differently
- Replication of chromosomal DNA begins at special sites called origins of replication, where the DNA duplex is unwound. Origins of replication tend to be full of A-T base pairs rather than G-C. What property of A-T pairing vs G-C pairing could explain this observation?
- DNA replication proceeds bidirectionally from the origin, with the enzyme helicase at the replication “fork” unwinding the DNA template in both directions away from the origin.
- DNA polymerase has to have an existing 3′-OH to add to, so it cannot start synthesizing a complementary strand from scratch. But RNA polymerase can start from scratch, so a new DNA strand starts with an RNA primer which is synthesized by primase. DNA polymerase adds new DNA nucleotides to the RNA primer to begin polymerizing DNA.
- DNA polymerase adds new nucleotides only at the 3′-OH end, so new strands grow 5′ –> 3′.
- DNA polymerase reads the template strand 3′ –> 5′, so the new DNA duplex is antiparallel.
- One daughter strand, called the “leading” strand, can be synthesized continuously.
- The other daughter strand, the “lagging” strand, has to be synthesized in short fragments, called “Okazaki” fragments.
- The RNA primers are replaced by DNA, and the Okazaki fragments are sealed by an enzyme called DNA ligase.
Go through this interactive animation; start with “Replication Fork,” then “Fork with Proteins,” “Concerted Replication,” and “Trombone Model”
https://www.courses.fas.harvard.edu/~biotext/animations/replication1.swf
You can visualize the difference in the leading and lagging strand here:

As the replication fork extends in one direction, the leading strand (top strands) can be synthesized continuously because it is proceeding in the 5′ to 3′ direction. The leading strand must be synthesized in short fragments as the DNA fork extends. This is because DNA synthesis must ALWAYS proceed in the 5′ to 3′ direction, but the lagging strand template is running the ‘wrong’ way for synthesis to occur in the direction of the replication fork (compare Time 1 and Time 2). Note both strands require RNA primers (yellow) to being DNA synthesis.
Here you can see that each side of the replication “bubble” or “eye” has a fork, and each fork has a leading and a lagging strand:

DNA replication animation:
DNAReplication
Hank Green’s take (see if you can spot the mistake on DNA structure):
Dr. Jung Choi’s video lecture on DNA replication (beginning at 14:52):
Other resources:
Molecular animation of the “trombone” model of DNA replication:
https://www.dnalc.org/resources/3d/04-mechanism-of-replication-advanced.html
Slide set to go with the lecture video above:
B1510_module4-5_DNA_Replication_2011